Research
Overview
Proteins are the body's "molecular machines" and are central in the countless processes that maintain all living organisms. A protein's function comes about as a direct result of the particular features of its three-dimensional structure. Thus, we need to know this structure in order to properly understand how a protein works, as well as to design drugs to modify the protein's function to treat a disease. The details of this structure are too small to be seen directly, even in the most highly magnified images, and so we use techniques such as nuclear magnetic resonance (NMR) to determine the structure.
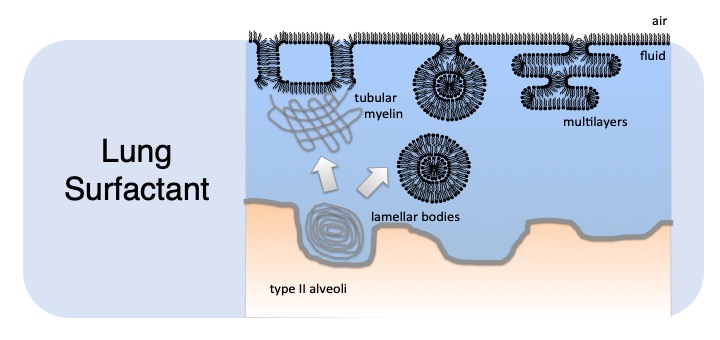
Lung surfactant (LS) is a mixture of lipids and proteins that coats the airspaces in our lungs. Without it, we can't breathe: premature babies are commonly born with deficiencies in LS (this condition is called RDS) and need to be given exogenous LS in order to survive. People of any age who are very ill can sustain damage to their lung surfactant (ARDS). Unfortunately for these people, giving them exogenous LS does not seem to help them very much, likely in part because whatever conditions present in their lungs that lead to ARDS in the first place, rapidly inactivate any additional LS given to them. Our work is aimed at understanding the molecular mechanisms of LS, particularly concerning the essential LS protein SP-B, as well as in designing "ARDS-resistant" peptides based on SP-B.
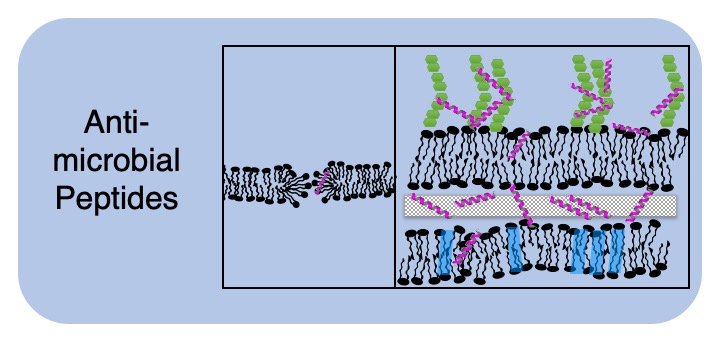
Although hundreds of peptide sequences with anti-bacterial/viral/fungal/tumor properties have been identified, we still have a very poor understanding of the molecular mechanisms by which they disrupt microbial cell function. Suitable antimicrobial peptides (AMPs) have much potential to address the current significant problem of bacterial (and other pathogen) resistance to small molecule antibiotics. In order to successfully develop antimicrobials as therapeutic drugs, it is important to understand how these molecules interact with bacteria. Much research has been carried out to characterize how AMPs interact with pathogen lipid membranes. We are currently focussed on how AMPs interact with other components of pathogens, such as cell envelope carbohydrates and intracellular molecular machines.
While biomolecules are most often characterized in dilute solution, their normal working environment is super-crowded, with concentrations up to 400 g/L! We are interested in how this crowding affects the structure and dynamics of proteins.