Extend single-locus ![]() multilocus
multilocus ![]() quantitative
models
quantitative
models
p2:2pq:q2
W0,W1,W2
Mendel & H-W Theorem
![]()
![]()
![]()
normal
distribution fitness
function Heritability
Quantitative genetics: Variation can be quantified
mean ![]() standard deviation:
standard deviation: ![]()
![]()
![]()
variance: ![]() 2
2
Quantitative variation follows "normal
distribution" (bell-curve) iff [read:
"if and only if"]
Multiple
loci involved
Each
locus
contributes equally (variance is additive)
Each locus acts independently:
interaction variance ( σ2GxE )
minimal
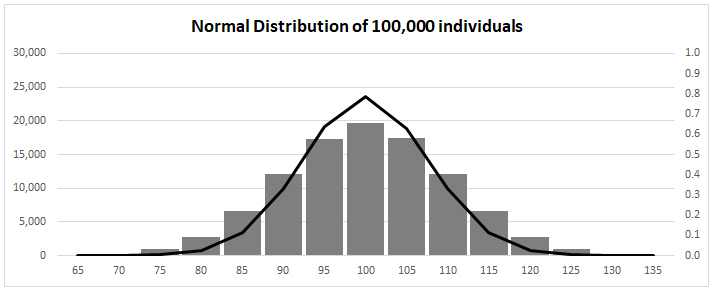
Normal Distribution for Mean 100 ![]() 10
10
Phenotypic variation has two
sources: genetic (σ2G)
& environmental (
phenotypic variance ![]() σ2P
= σ2G
+
σ2P
= σ2G
+
additive variance ![]() σ2A
= σ2G
+
σ2A
= σ2G
+
heritability
![]() h2 = σ2G/ σ2A
= σ2G
/ (σ2G
+
h2 = σ2G/ σ2A
= σ2G
/ (σ2G
+
h2 : "Heritability
in the narrow sense": ignores ![]() GxE2 = interaction
variance:
GxE2 = interaction
variance:
Identical genotypes produce different phenotypes
in different environments.
Ex.: same breed of cows produces different milk
yield on different feed
Norm of
Reaction for differential expression within & between
environments
Artificial
breeding indicates that organismal phenotypic
variation is highly heritable
ex.:
Darwin's pigeon breeding
experiments
Artificial
selection on agricultural species
Commercially
useful
traits
improved by selective breeding
IQ scores in Homo:
h2 ~ 0.7
But: IQ scores improve with education:
Offspring /
Mid-parent correlation
Fitness
function expresses relationship between
genotype & fitness
A continuous
variable over range of genotypes, rather than discrete
values for W0, W1, & W2
Most traits variable &
heritable
Many traits do
respond to 'artificial'
selection (h2 = 0.5 ~
0.9)
Many traits should
respond to 'natural'
selection
To demonstrate & measure
Natural Selection in Nature,
Show experimentally
that heritable
variation has consequences for
fitness
What
happens
to
a
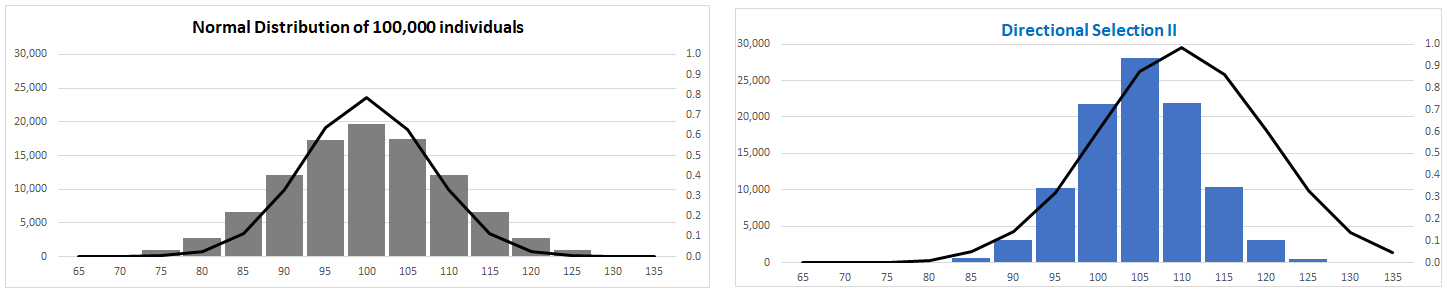
normal distribution under Selection?
Compare distributions before [left] and after [right]
selection
Directional
Selection
(I) Fitness function has constant
slope:
Trait mean shifted towards
one extreme of phenotype
trait variance changed [skewed to
right]
(II) Fitness function normally
distributed
Trait mean shifted: variance unaffected

In
single-locus models, limit of selection is
Elimination
of variation by fixation of favored allele
Clinal
variation of B allele in human ABO
system
In quantitative
models, rate limited by
substitutional genetic load:
Fitness "cost" (lost
reproductive potential) to replace disadvantageous
allele
"Soft" selection
Mortality is density-dependent
N(after) ~ N(before)
Survivorship proportional to fitness up to K: more
realistic
Selection affects recruitment to next generation
Ex.: In AS system, deaths from malaria or
sickle-cell are continuously "replaced"
N continually "topped up" to K
Darwin's Finch (Geospiza fortis) adapts to
drought:
larger birds survive because of changes in seed size & hardness
Developmental canalization
limits extent of directional selection
Systems
are
controlled
by
multiple epistatic loci:
difficult to select on all loci simultaneously
Organisms have developmental limits:
e.g. size cannot increase indefinitely
Johanssen bean
experiment exhausted genetic variation within
lineage
(2) Stabilizing
Selection (AKA truncation selection)
Fitness
function has "peak"
Variance reduced around constant trait mean at
optimal phenotype,
or tails of
distribution eliminated (truncated)
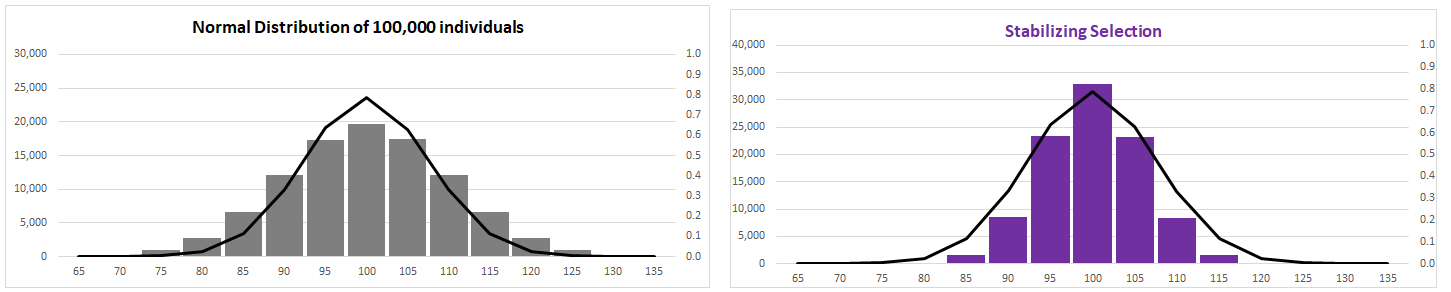
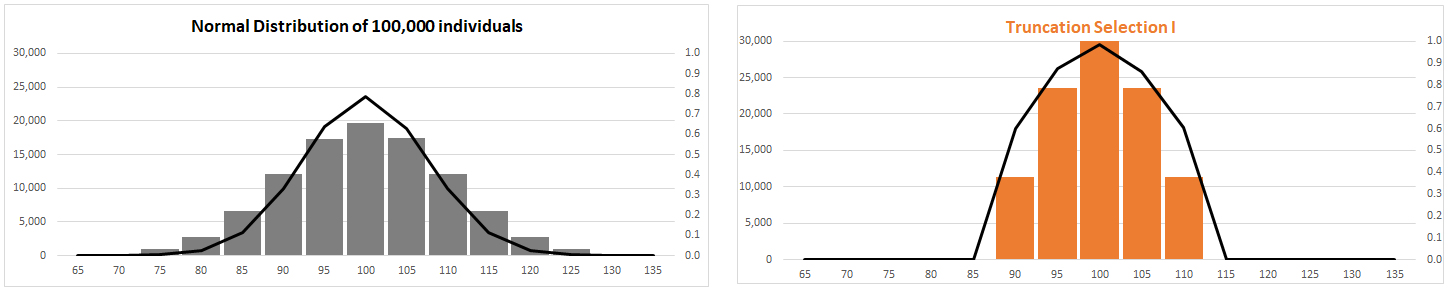
Limits: elimination of variant alleles
or, 'weeding out' of disadvantageous variants
homozygosity at multiple loci:
difficult iff variance due to recessive alleles
inbreeding depression: loss
of 'somatic vigor' in inbred lines
Ex.: Birth
weight in Homo
Modal
birth weight has optimum survival
Implications for future human
evolution ?
(3) Diversifying
Selection (two kinds)
There
is a lot of variation: can natural selection explain it?
No single optimal from:
trait variance increases
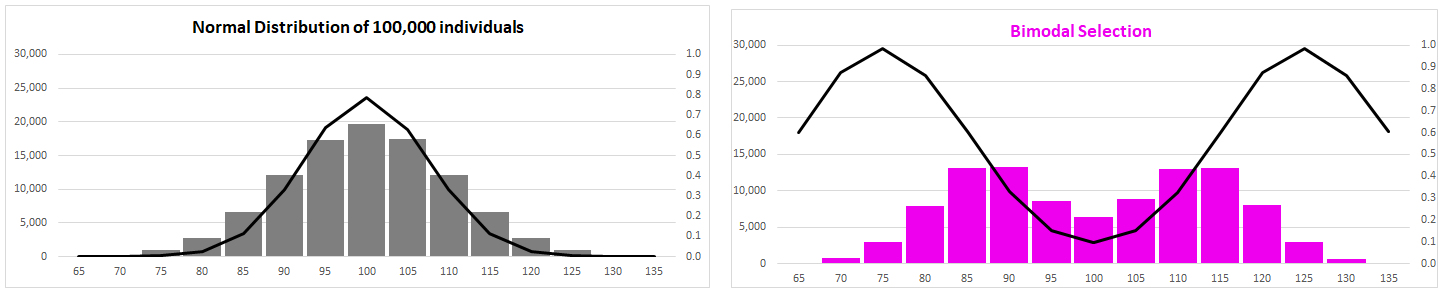
polymorphic:
variation maintained within populations
Ex.: Genetic variation in corn
snakes, tomatoes, bell peppers, snails
Ex.: shell
patterns in Cepaea snails
combinations of dark / light, banded / unbanded
shells varies with substrate
polytypic:
variation distributed among populations
Ex.: Clinal variation in Cepaea
snails
patterns
of banded / unbanded shells vary over short distances
Müllerian mimicry:
Ex.: Heliconiusspp.
butterflies exchange alleles by hybridization
Distasteful models converge on
each other,
Different
combinations evolve in different parts of range
Limits:
segregational genetic load:
Fitness "cost"
(loss of reproductive potential) from production of less
fit homozygotes
Overdominance: heterozygotes
have superior fitness
because
different
alleles
favored in different environments
Heterodimers:
multimeric enzymes with polypeptides from different
alleles
often show wider substrate specificity, kinetic properties (Vmax
& KM)
Myoglobin in
diving mammals
Heterosis: heterozygosity at
multiple loci correlated with overall fitness
Ex.: correlation between phenotype
& He: antler points in deer
(Odocoileus)
Ex.: Hybrid
vigor: crossbreeding
of inbred lines in maize (Zea)
improves vigor in F1
Maintaining polymorphic multi-locus phenotypes by Natural Selection
Alternative phenotypes favored in different environments
Batesian mimicry:
'Tasty' mimics converge on 'distasteful' models
Ex.: Viceroy butterflies (Limenitis) converge on Monarch
(Papilio) butterflies
Mimic remains
rare wrt model
Müllerian mimicry:
Distasteful models converge on each other,
Different combinations evolve in different
parts of range
Ex.: Heliconius
spp. butterflies exchange alleles by hybridization
Mertensian mimicry:
aposematic (warning)
coloration discourages predators
Ex.: non-venomous scarlet king snakes
mimic
venomous coral snakes with black / red / yellow
pattern
Frequency-dependent selection:
apostatic predation: Turdus
thrush predation on Cepaea snails
'search image' changes when prey type becomes rare
Sexual Selection (Darwin 1871):
'Exaggerated'
ornamentation disadvantageous to individual survival
phenotype
but favored in competition for mates (reproductive phenotype)
Sexual dimorphism in birds
& mammals
Antlers in deer (Cervidae)
used in male-male
combat
Runaway sexual selection
Females
choose
males
on
basis of secondary sex ornamentation
Offspring have exaggerated trait (males) & preference
for trait (females)
 selection reinforces trait &
preference for trait simultaneously
selection reinforces trait &
preference for trait simultaneously
New phenotype spreads
rapidly in population
(B) Disruptive
selection
Fitness function is a valley
Trait variance increases (like balancing selection), but
polymorphism unstable
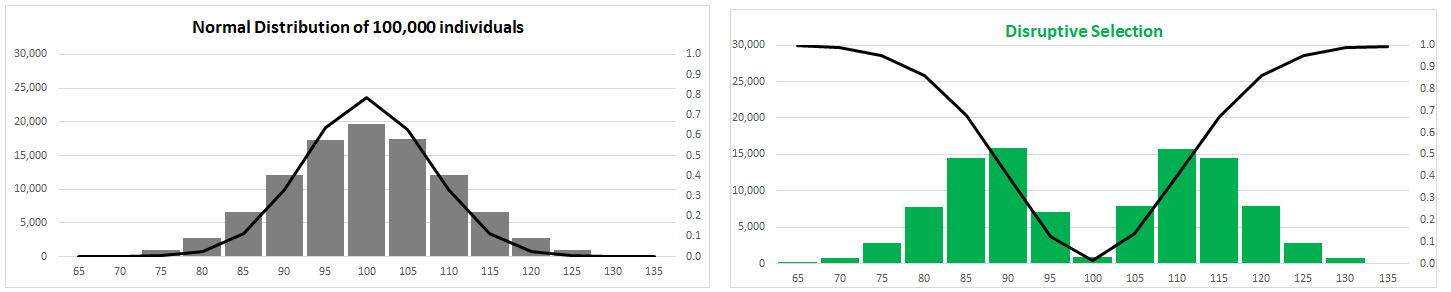
Polymorphism can usually be maintained only temporarily:
One
of
the
phenotypes
will out-compete the other
unless different phenotypes choose different niches (Ludwig
Effect)
then this becomes Balancing Selection
or frequency
selection occurs
Scutellar
bristles in Drosophila
Selection for 'high #' versus 'low #' lines
=> 'pseudo-populations'
with reduced inter-fertility
Might
disruptive
selection
contribute
to speciation?
Darwinian natural selection defined
as
differential
survival & reproduction of individuals:
Can
selection operate on more inclusive biological
units?
Group
Selection controversy: cf. Kropotkin
(1902) "Mutual Aid"
Kin
(Interdemic) Selection
![]() Differential survival & reproduction
of related (kin) groups (extended families)
Differential survival & reproduction
of related (kin) groups (extended families)
Related
individuals share alleles: r =
coefficient of relationship [derivation]
offspring
&
parents
related
by r = 0.50 [They share
half their alleles]
full-sibs
"
"
r = 0.50
half-sibs
"
"
r = 0.25
first-cousins
"
"
r = 0.125
Inclusive fitness (WI) of
phenotype for individual i
= direct fitness of i + indirect
fitness of relatives j,k,l,...
WI = ai + ![]() (rij)(bij) summed over all relatives
j,k,l,...
(rij)(bij) summed over all relatives
j,k,l,...
where: ai = fitness of i
due to own phenotype
bij = fitness of j
due to i's phenotype
rij =
coefficient of relationship of i &
j
If i & j are unrelated
warn: Windividual
= 0.0 + (0.0)(1.0) = 0.0
don't warn: Windividual = 1.0 +
(0.0)(0.0) = 1.0
 Such behaviors should not
evolve among unrelated individuals
Such behaviors should not
evolve among unrelated individuals
What
is
the
fitness
value in a kin group?
Wbrothers = 0.0
+ [2](0.5)(1.0)] = 1.0
Wcousins =
0.0 + [8][(0.125)(1.0)] = 1.0
 Such behaviors can evolve among related
individuals in (extended) family groups
Such behaviors can evolve among related
individuals in (extended) family groups
JBS Haldane (1892 -
1964):
"I would lay down my life for two
brothers or eight cousins."
Humans evolved
in small kin groups, function today in large, ~unrelated
groups:
do evolved Primate behaviors persist?
Parenting behavior:
'Broken wing'
display in mother birds
Mother sacrifices herself for
(at least two) offspring
[See also active
defense of offspring] [Warning:
language!]
Altruistic behavior (![]() "unselfish concern
for others")
"unselfish concern
for others")
Alarm
calls in ground squirrels (Spermophilus)
females
warn
more
in
related groups
Can
behavior
help
unrelated individuals evolve?
Eusocial insects (Hymenoptera, Isoptera)
Haplodiploidy: females diploid, male drones
haploid
Females
workers
sterile
(Wi = 0): what selective advantage?
related
to
queen
by 1/2
related to sisters by 3/4
 Care for sisters, don't have offspring
Care for sisters, don't have offspring
[NB:
'Queen' Bee mates with multiple drones: workers
related to half-sisters only by 1/2]
Text material © 2025 by Steven M. Carr