Segregational Genetic Load as a limit to genetic variance
In older discussions as to how
much genetic variation can be maintained in natural
populations, the Balanced School argued that high
levels of heterozygosity could be maintained by overdominant
selection at multiple loci, if alternative alleles at
a locus were advantageous under different circumstances
(e.g., colder & warmer seasons, wetter & drier
environments, or kidney & brain tissue types). The
classical example is balancing selection for sickle-cell
anemia polymorphism: the example is difficult to replicate.
The counter-argument of the Classical
School was that, in such a model loss of fitness
from maintenance of homozygotes with
sub-optimal fitness would severely limit the number of
heterozygous loci possible. This loss of fitness is called segregational load, because
it arises from segregation of the A &
B alleles at a locus into gametes as AA, AB
& BB genotypes, with AB as having
the greatest fitness.
Recall that the equilibrium frequency for an
allele B with a heterozygote
advantage is
![]() = s1
/ (s1 + s2)
= s1
/ (s1 + s2)
and the companion formula for A is
![]() = s2
/ (s1 + s2)
= s2
/ (s1 + s2)
If the relative fitness values of the AA,
AB, & BB genotypes are (1 - s1),
1, & (1 - s2),
respectively, the combined reduction in fitness
due to maintenance of AA and BB is
(s1p2
+ s2q2)
Substituting the equilibria ![]() and
and
![]() into this formula as the squares of p
and q,
into this formula as the squares of p
and q,
(s1)(s2)2 / (s1 + s2)2 + (s2)(s1)2 / (s1 + s2)2
[(s1)(s2)2
+ (s2)(s1)2]
/ (s1 +
s2)2
(s1)(s2)(s1
+ s2) / (s1
+ s2)2
Segregational load = (s1)(s2) / (s1
+ s2)
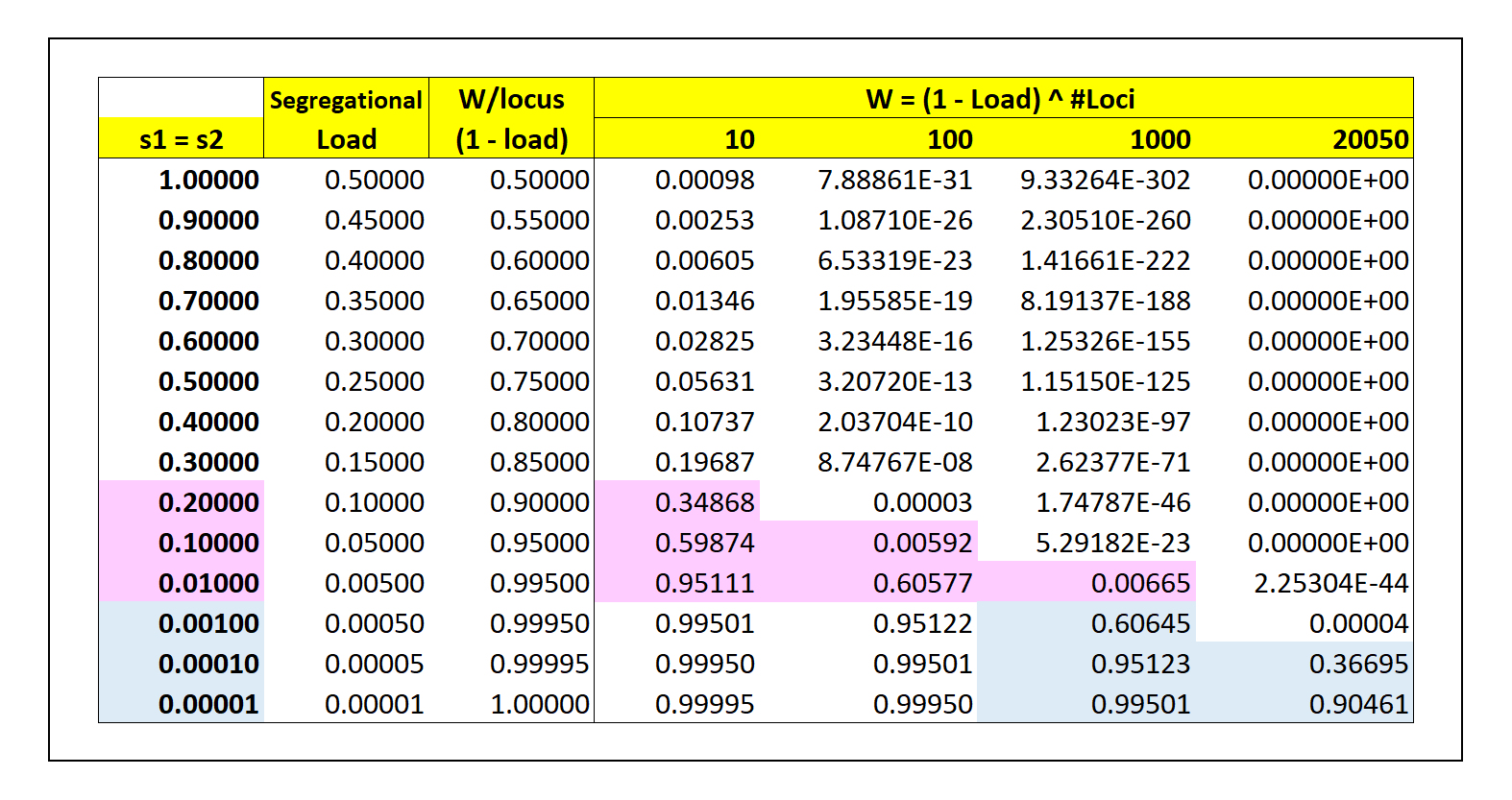
How many variable loci can be maintained, at what cost
with respect to fitness? The table above shows, for the
simple case of s1
= s2, the reduction in fitness
per locus, and the residual Fitness for
various numbers of heterozygous loci. (Note that in
this case, Segregational Load = s /
2).
The Classical argument
is that for reasonable observed values of s1 = s2
~ 0.1, 10 such loci would reduce
fitness to ~60%, the maximum tolerable.
Therefore, overdominant selection cannot maintain
high heterozygosity at more than a very few loci:
the vast majority of loci must be
homozygous.
The Balanced
argument is that for possible values
of s1 = s2
~ 0.001, 1000 or more heterozygous
loci could be maintained at ~60%
fitness as above. Populations can be highly
variable. However, selection coefficients on this
order are effectively impossible to measure
accurately.
Data from protein
electrophoresis and now from DNA
sequencing show that multiple thousands of
loci are polymorphic, apparently defeating the
Classical position. There is however a rejoinder: if
alternative alleles at a locus are neutral (s1 = s2
= 0.0) or nearly
neutral (s1
= s2
< 0.00001)
in their effect on
fitness,
an effect that could not be measurable
experimentally, 1000's of loci could be heterozygous
without serious loss of fitness. At the limit as
shown in the table, heterozygosity at all
20,050 protein-coding loci in the human
genome could be maintained at a cumulative cost of
10% reduction in overall fitness.
The Classical
/ Balanced argument then shifts
from a question of the value of Hobs
to a question of the value of the selection
coefficient s of any
observed allelic variant on fitness. The Classical / Balanced argument
pirouettes to the Selectionist
/ Neutralist argument.
It also revives and reinvigorates the argument that
it is not protein variation per se that is
important in evolution, but other aspects of
genomics, such as gene regulation. It
remains true that many traits of adaptive or medical
importance are due to single-locus variation, such
as hemoglobin polymorphisms in deep-diving vs
terrestrial mammals, or sickle-cell alleles.