Extend single-locus ![]() multilocus
multilocus ![]() quantitative
models
quantitative
models
p2:2pq:q2
W0,W1,W2
Mendel & H-W Theorem
![]()
![]()
![]()
normal
distribution fitness
function Heritability
Quantitative genetics: Variation can be quantified
mean ![]() standard deviation:
standard deviation: ![]()
![]()
![]()
variance: ![]() 2
2
Quantitative variation follows "normal
distribution" (bell-curve) iff [read:
"if and only if"]
Multiple
loci involved
Each
locus
contributes equally (variance is additive)
Each locus acts independently:
interaction variance ( σ2GxE )
minimal
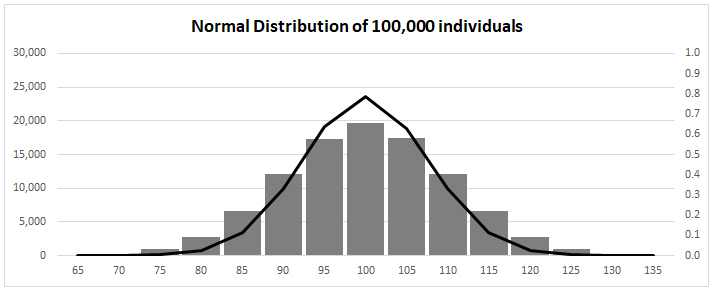
Normal Distribution for Mean 100 ![]() 10
10
Phenotypic variation has two
sources: genetic (σ2G)
& environmental (
phenotypic variance ![]() σ2P
= σ2G
+
σ2P
= σ2G
+
additive variance ![]() σ2A
= σ2G
+
σ2A
= σ2G
+
heritability
![]() h2 = σ2G/ σ2A
= σ2G
/ (σ2G
+
h2 = σ2G/ σ2A
= σ2G
/ (σ2G
+
h2 :
"Heritability in the narrow sense": ignores ![]() GxE2 = interaction
variance:
GxE2 = interaction
variance:
Identical genotypes produce different phenotypes
in different environments.
Ex.: same breed of cows produces different milk
yield on different feed
Norm of
Reaction for differential expression within & between
environments
Artificial
breeding indicates that organismal phenotypic
variation is highly heritable
ex.:
Darwin's pigeon breeding
experiments
Artificial
selection on agricultural species
Commercially
useful
traits
improved by selective breeding
IQ scores in Homo:
h2 ~ 0.7
But: IQ scores improve with education:
Offspring /
Mid-parent correlation
Fitness
function expresses relationship between
genotype & fitness
A continuous
variable over range of genotypes, rather than discrete
values for W0, W1, & W2
Most traits variable &
heritable
Many traits do
respond to 'artificial'
selection (h2 = 0.5 ~
0.9)
Many traits should
respond to 'natural'
selection
To demonstrate & measure
Natural Selection in Nature,
Show experimentally
that heritable
variation has consequences for
fitness
What
happens
to
a
normal distribution under Selection?
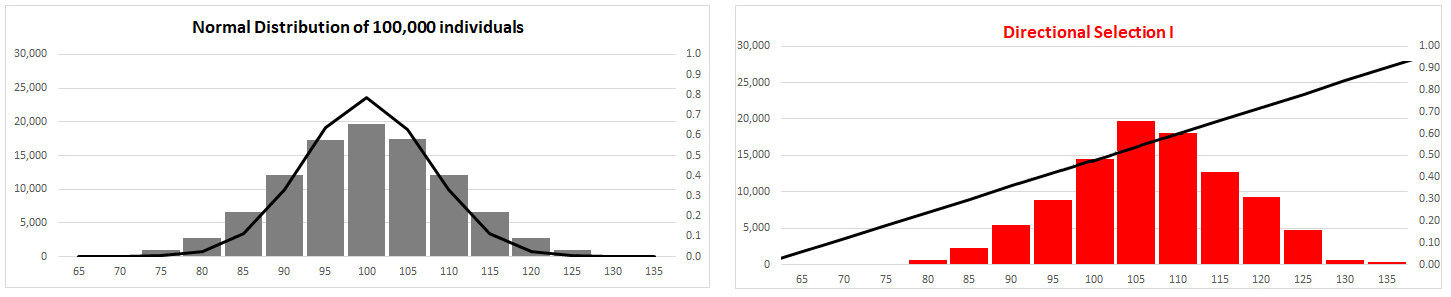
Compare distributions before [left] and after [right]
selection
Directional
Selection
(I) Fitness function has constant
slope:
Trait mean shifted towards
one extreme of phenotype
trait variance changed [skewed to
right]
NB: Fitness function resembles that for favored
single allele
Variance corrected by HWE each generation
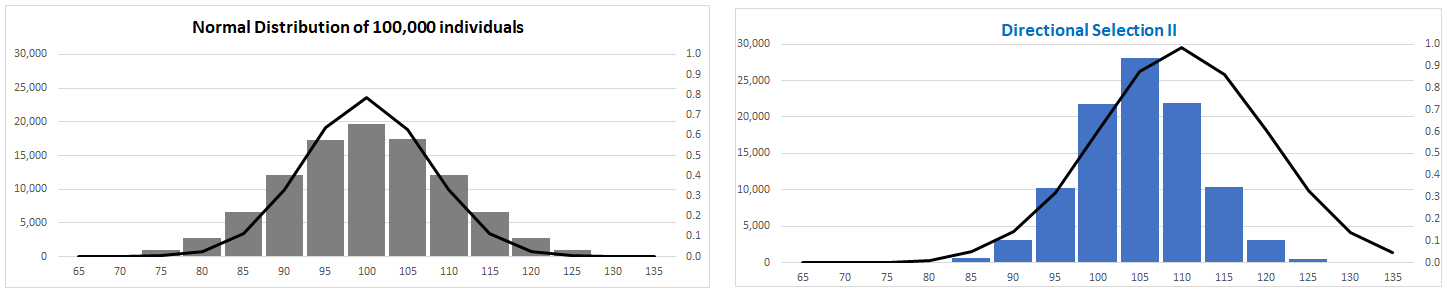
(II)
Fitness function normally distributed
Trait mean shifted gradually: variance unaffected
NB: Fitness function resembles that for
heterozygote superiority:
What's different?
HOMEWORK:
Compare and contrast predictions of single-locus versus multi-locus
selection.
In
single-locus models, limit of selection is
Elimination
of variation by fixation of favored allele
Clinal
variation of B allele in human ABO
system: f(B) declines East to West
In quantitative
models, rate limited by
substitutional genetic load:
Fitness "cost" (lost
reproductive potential) to replace disadvantageous
allele
"Soft" selection
Mortality is density-dependent
N(after) ~ N(before)
Survivorship proportional to fitness up to K: more
realistic
Selection affects recruitment to next generation
Ex.: In AS system, deaths from malaria or
sickle-cell are continuously "replaced"
N continually "topped up" to K
Darwin's Finch (Geospiza fortis) adapts to
drought:
larger birds survive because of changes in seed size & hardness
Developmental canalization
limits extent of directional selection
Systems
are
controlled
by
multiple epistatic loci:
difficult to select on all loci simultaneously
Organisms have developmental limits:
e.g. size cannot increase indefinitely
Johanssen bean
experiment exhausted genetic variation within
lineage
(2) Stabilizing
Selection (AKA truncation selection)
Fitness
function has "peak"
Variance reduced around constant trait mean at
optimal phenotype,
or tails of
distribution eliminated (truncated)
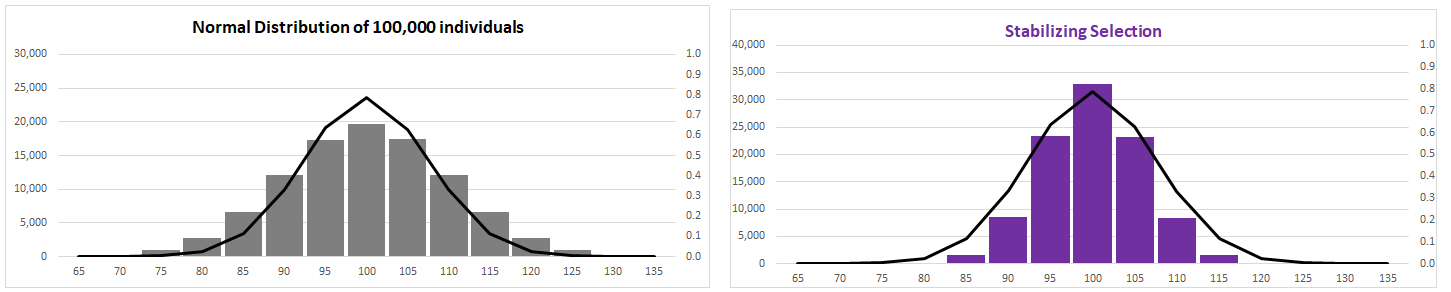
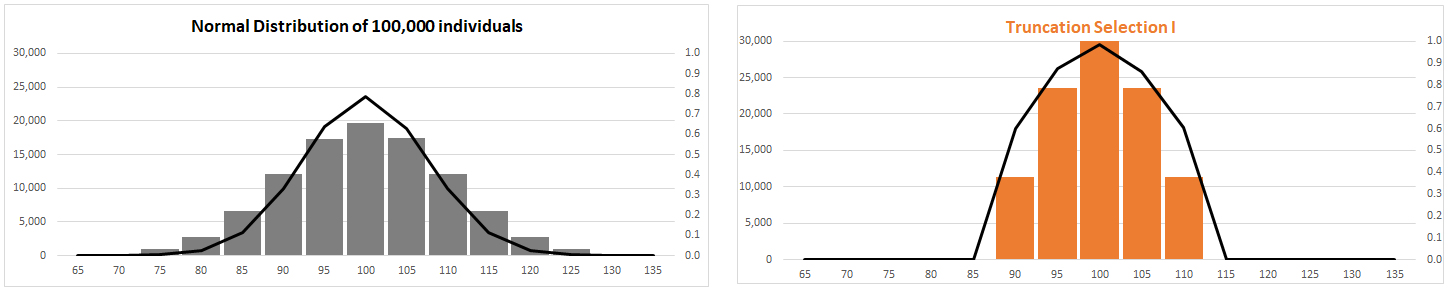
Limits: elimination of variant alleles
or, 'weeding out' of disadvantageous variants
homozygosity at multiple loci:
difficult iff variance due to recessive alleles
inbreeding depression: loss
of 'somatic vigor' in inbred lines
Ex.: Birth
weight in Homo
Modal
birth weight has optimum survival
Implications for future human
evolution ?
(3) Diversifying
Selection (two kinds)
There
is a lot of variation: can natural selection explain it?
No single optimal from:
trait variance increases
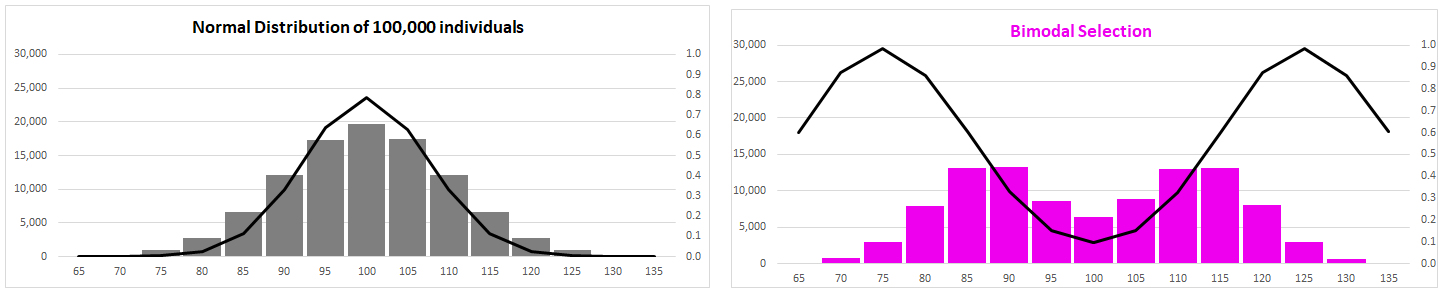
polymorphic:
variation maintained within populations
Ex.: Genetic variation in corn
snakes, tomatoes, bell peppers, snails
Ex.: shell
patterns in Cepaea snails
combinations of dark / light, banded / unbanded
shells varies with substrate
polytypic:
variation distributed among populations
Ex.: Clinal variation in Cepaea
snails
patterns
of banded / unbanded shells vary over short distances
Müllerian mimicry:
Ex.: Heliconiusspp.
butterflies exchange alleles by hybridization
Distasteful models converge on
each other,
Different
combinations evolve in different parts of range
Text material © 2025 by Steven M. Carr