

I am interested in
patterns and processes of molecular
phylogeography and evolution within and among species and
natural populations of vertebrate animals, and novel genomic biotechnology
approaches to these questions. My investigations include:
(1) Mitogenomic
Phylogeography of the pre- & post-glacial Nearctic &
North Atlantic, including Atlantic Cod, Wolffish, Harp Seals,
and Newfoundland Caribou
(2) Genomic diversity &
origins of ancient and modern peoples of Newfoundland
(3) Phylogeography of "Species At Risk" of
extinction, including the occasional "sea monster"
For further details and other
projects, please see the PDF MSS below, and (or)
contact Steve at scarr@mun.ca.
My laboratory has
taken a mitogenomic approach to the interrelated problems
of evolutionary and population genetics and systematic biology.
Analyses based on complete maternally-inherited mitochondrial
DNA genomes provide well-resolved, highly-corroborated
molecular phylogenies within and among species that can generate
and test hypotheses about morphological, biogeographic, and
behavioral evolution. Phylogeographic analysis
[the analysis of genetic relationships in their geographic
context] of completely-resolved intraspecific gene trees, based on
full-length mtDNA genomes,
provides the detailed historical information and necessary
statistical power to evaluate vicariance and dispersal phenomena
at scales of interest to fisheries managers and population
biologists. I have combined this with novel analytical methods,
including a Monte Carlo simulation test of population
structure, and a non-parametric Kolmogorov-Smirnoff test
of coalescent offsets.
I am
cross-appointed to the Department of Computer Science,
where my interests are in Computational Biology of DNA
sequences and Next-Generation DNA data
signal analysis.



S Royston & SM Carr. 2016a. Conservation genetics of high-arctic gulls at risk. I. Diversity in the mtDNA Control Region of circumpolar populations of the Endangered Ivory Gull (Pagophila eburnea). Mitochondrial DNA 27, 3995-3999. [PDF]
SM Carr, AT Duggan, GB Stenson, & HD Marshall. 2015. Quantitative phylogenomics of within-species mitogenome variation: Monte Carlo and non-parametric analysis of phylogeographic structure among discrete transatlantic breeding areas of Harp Seals (Pagophilus groenlandicus). PLoS ONE,10(8): e0134207. [PDF] s
s SM Carr, HT Wareham & Craig D.
2014. A web
application for generation of DNA sequence exemplars with
open and closed reading frames in genetics and bioinformatics
education. CBE – Life Sciences Education 13,
373-374. [PDF]
SM Carr, D Craig,
& HT Wareham. 2014 An algorithmic and computational
approach to Open Reading Frames in short dsDNA sequences:
Evaluation of “Carr’s Conjecture.” Pp. 37-48 in Proceedings
of the 14th International Conference on Bioinformatics
& Computational Biology. [H. Arabnia et al.,
eds.]. [PDF]
SM Carr. 2013. Composite and
complementary DNA. Lab Business, Nov / Dec 2013. [PDF]
SM Carr. 2013. "Known Knowns,
Known Unknowns, & Unknown Unknowns": Computational
science challenges for analysis of multidimensional DNA matrices
in evolutionary & population genomics. Pp. 508-512 in Proceedings
of the 13th International Conference on Bioinformatics &
Computational Biology (H. Arabnia, ed.]. [PDF]
AM Pope, SM Carr, KN Smith, & HD
Marshall. 2011. Mitogenomic and
microsatellite variation in descendants of the founder
population of Newfoundland: high genetic diversity in an
historically isolated population. Genome 54,110-119. [PDF]
SM Carr, AT Duggan, & HD Marshall. 2009. Iterative DNA sequencing on microarrays: a high-throughput NextGen technology for ecological and evolutionary mitogenomics. Laboratory Focus 13, 8-12. [PDF]
HD Marshall, MW Coulson, & SM Carr. 2008. Near neutrality, rate heterogeneity, and linkage govern mitochondrial genome evolution in Atlantic Cod (Gadus morhua) and other gadine fish. Molecular Biology & Evolution 26, 579-589. [PDF]
SM Carr & HD Marshall. 2008. Phylogeographic analysis of complete mtDNA genomes from Walleye pollock (Gadus chalcogrammus Pallas, 1811) shows an ancient origin of genetic biodiversity. Mitochondrial DNA 19, 490-496. [PDF]
SM Carr & HD Marshall. 2008. Intraspecific phylogeographic
genomics from multiple complete mtDNA genomes in Atlantic Cod (Gadus morhua): Origins of the "Codmother," trans-Atlantic vicariance, and
mid-glacial population expansion. Genetics 108, 381-389. [PDF]
SM Carr, HD Marshall, AT Duggan, SMC Flynn, KA Johnstone, AM Pope, & CD Wilkerson. 2008. Phylogeographic genomics of mitochondrial DNA: patterns of intraspecific evolution and a multi-species, microarray-based DNA sequencing strategy for biodiversity studies. Comparative Biochemistry and Physiology, D: Genomics and Proteomics 3,1-11. [PDF]
SJ Moore, DJ Buckley, A MacMillan, HD Marshall, L Steele, P Ray, Z Nawaz, M Frecker, SM Carr, E Ives, & PS Parfrey. 2008. The clinical and genetic epidemiology of neuronal ceroid lipofuscinosis in Newfoundland. Clinical Genetics 74, 213-222. [PDF]
SMC Flynn &SM Carr. 2007. Interspecies hybridization on DNA
resequencing microarrays: efficiency of sequence recovery and
accuracy of SNP detection in human, ape, and codfish
mitochondrial DNA genomes sequenced on a human-specific
MitoChip. BMC Genomics
8, 339. [PDF]
KA Johnstone, HD Marshall, & SM
Carr. 2007. Biodiversity
genomics for Species At Risk: patterns of DNA sequence variation
within and among complete mitochondrial DNA genomes of three
species of Wolffish (Anarhichas spp.). Canadian
Journal of Zoology 85,151-158. [PDF]
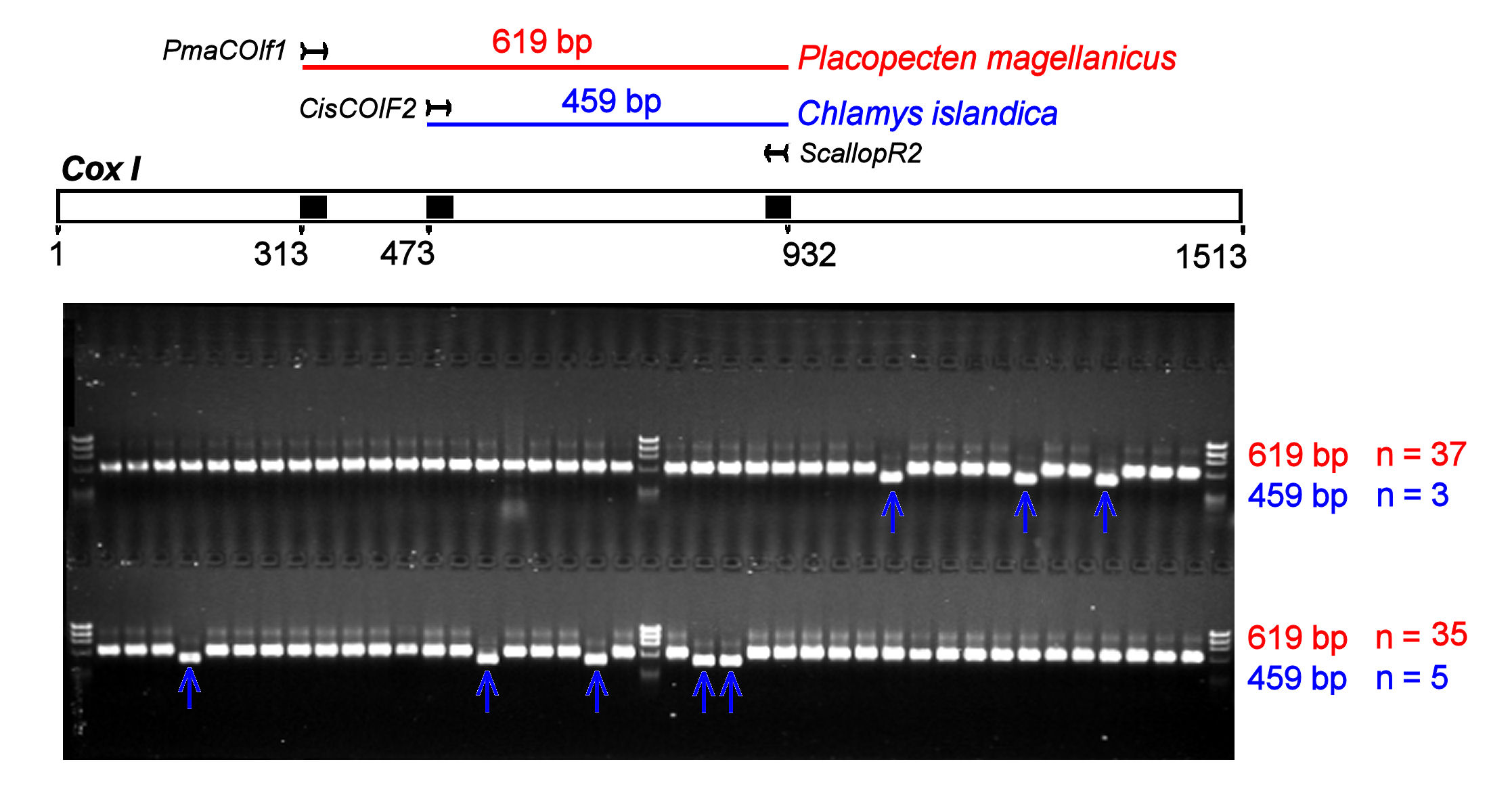
HD Marshall, KA Johnstone, & SM Carr. 2006. Species-specific oligonucleotides and
multiplex PCR for forensic discrimination of two species of
scallops, Placopecten
magellanicus and Chlamys
islandica. Forensic
Science International 167,1-7. [PDF]
MW Coulson, HD Marshall, P Pepin & SM Carr. 2006. Mitochondrial phylogeographic genomics of gadine fish: Implications for taxonomy and biogeographic origins. Genome 49,1115-1130. [PDF]
SM Carr, HD Marshall, KA Johnstone, LM Pynn, & GB Stenson. 2002. How To Tell a Sea Monster: Molecular Discrimination of Large Marine Animals of the North Atlantic. The Biological Bulletin 202,1-5. [PDF]
EA Perry, GB Stenson, SE Bartlett, WS Davidson, & SM Carr. 2000. DNA sequence analysis identifies genetically distinguishable populations of harp seals (Pagophilus groenlandicus) in the northwest and northeast Atlantic. Marine Biology 137, 53- 58. [PDF]
SM Carr, DGS Kivlichan, P Pepin &
DC Crutcher. 1999. Molecular
phylogeny of gadid fishes: implications for the biogeographic
origins of Pacific species. Canadian Journal of
Zoology 77,19-26. [PDF]
SM Carr & DC Crutcher. 1998. Population genetic structure in
Atlantic Cod (Gadus morhua) from the North Atlantic and Barents Sea: contrasting or
concordant patterns in mtDNA sequence and microsatellite data?
Pp. 91-103 In The Implications of Localized Fishery Stocks (I.
Hunt von Herbing, I. Kornfield, M. Tupper, and J. Wilson. eds.).
Northeast Regional Agricultural Engineering Service, Ithaca, New
York.[ PDF]
ML Vis, SM Carr, WR Bowering, &
WS Davidson. 1997.Greenland Halibut (Reinhardtius
hippoglossoides) in the North Atlantic are genetically
homogeneous Canadian Journal of Fisheries & Aquatic
Sciences 53, 1813-1821. [PDF]
SM Carr & EA Perry.
1997. Intra- and
interfamilial systematic relationships of phocid seals as
indicated by mitochondrial DNA sequences. Pp. 277-290
in A. E. Dizon et al. (eds). Molecular Genetics of Marine
Mammals. Special Publication No.3 of the Society for
Marine Mammalogy, Lawrence KS.
EA Perry, SM Carr, SE
Bartlett, & WS Davidson.
1995. A phylogenetic
perspective on the evolution of reproductive behavior in
pagophilic seals of the Northwest Atlantic as indicated by
mitochondrial DNA sequences. Journal of Mammalogy 76,
22-31. [ PDF]
SM Carr, SW Ballinger, JN Derr, LH
Blankenship, & JW Bickham. 1986. Mitochondrial DNA analysis of hybridization between
sympatric white-tailed deer and mule deer in west Texas.
Proceedings of the National
Academy of Science (USA) 83, 9576-9580 [PDF]
AC Wilson, RL Cann, SM Carr, M
George, UB Gyllensten, KM Helm-Bychowski, RG Higuchi, SR Palumbi,
EM Prager, RD Sage, & M Stoneking. 1985. Mitochondrial DNA
and two perspectives on evolution. Biological Journal of
the Linnaen Society 26, 375-400 [PDF]

Bio2250
- Principles of Genetics
Bio2900 -
Principles of Evolution & Systematics
Bio4241 -
Advanced Genetics
Bio4250 - Evolutionary Genomics
Bio4270 - History of Biology
Bio4900 - Fundamentals of Genetic
Biotechnology
Med6392 - Human
Population Genetics