The Polymerase Chain Reaction (PCR)

The Polymerase Chain Reaction (PCR)
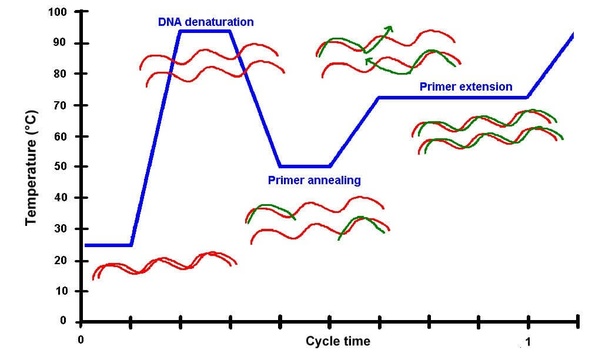
PCR begins with a mixture containing a dsDNA template, a pair of short ssDNA oligonucleotide primers, a pool of the four dNTPs, and a heat-resistant DNA polymerase, Taq Enzyme. The reaction is carried out in a computer-regulated heating block, a thermal cycler, which permits rapid, controlled heating & cooling. The primers are chosen so that they are base-complementary to opposite ends of either strand of a several hundred base pair region of DNA containing the gene region of interest: PCR thus requires some prior knowledge of the gene. In the case of molecular systematic studies, many so-called "Universal Primers" target regions that are identical across a wide range of taxa.
The reaction is first heated to 95oC to melt (denaturation) the dsDNA into separate strands. The reaction is then cooled to ~50oC, at which temperature the primers will find base-complementary regions in the ssDNA, to which they will stick (annealing). The reaction is finally heated to 72oC, at which temperature the Taq enzyme replicates the primed ssDNA (extension). At the end of one cycle, the region between the two primers has been copied once, producing two copies of the original gene region. [This is slightly oversimplified: see details].
The key
innovation that made PCR practical was the use of a
heat-resistant polymerase, so that the reaction can be
repeated continuously without addition of more enzyme. Each
cycle doubles the copy number of the amplified gene: ten
cycles ideally produces 2 ![]() 4
4 ![]() 8
8 ![]() 16
16 ![]() 32
32 ![]() 64
64 ![]() 128
128 ![]() 256
256 ![]() 512
512 ![]() 1,024
(210) copies.
Thus, 30 cycles yields a (210x3)
= 109-fold
amplification. This produces a sufficient quantity of the
gene region of interest for direct analysis, for example by DNA
sequencing.
1,024
(210) copies.
Thus, 30 cycles yields a (210x3)
= 109-fold
amplification. This produces a sufficient quantity of the
gene region of interest for direct analysis, for example by DNA
sequencing.