LOD (Logarithm Of Odds) score analysis II:
Some mathematical considerations
LOD score analysis is
used to estimate whether the observed degree of concordance of a genetic marker
with a trait of interest indicates genetic
linkage between the two. LOD
analysis is typically used in Genome-Wide Association Studies (GWAS) to map a trait of
interest to a particular chromosomal region. LOD score analysis
requires a fully-mapped genome with many thousands of genetic
markers on all chromosomes. and is complicated by many factors
that are built into the mathematical model. Moving beyond a simple
model requires some appreciation of of the math.
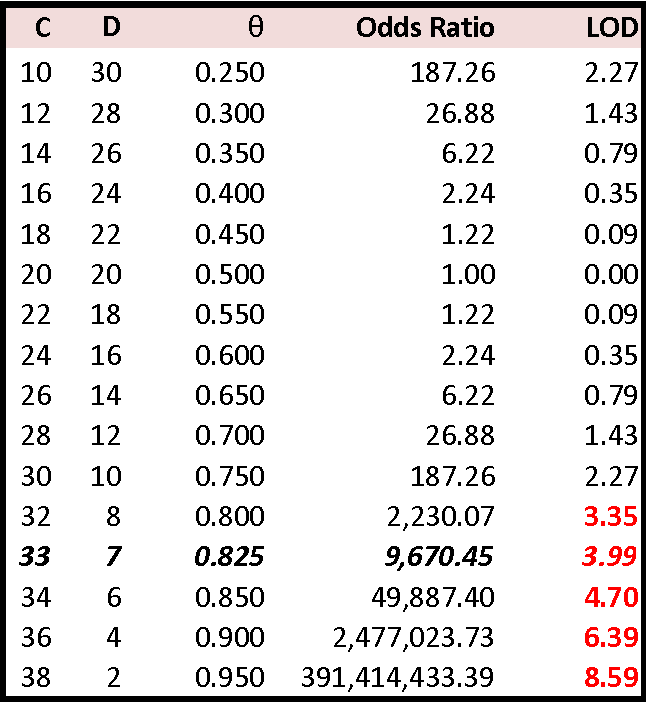
The conventional approach to a single-classification problem (concordance or non-concordance, with either outcome of equal probability) would be a Chi-square test. Significant concordance in the above table occurs at 26:14, for which X2 = 3.60 and p ~ 0.05. However, where multiple locus comparisons are done simultaneously, about one concordance test in 20 is expected to be show a significant score, simply by chance. Further, the Chi-square test is a parametric test that assumes a certain distribution of linkage relationships, which may not be so. Thus the preferred statistically approach is an odds ratio.
For each marker, the Odds Ratio = [ (θC)(1-θ)D ] / 0.5C+D , where C is the number of concordant pairs, D the number of discordant pairs (thus N = C+D), and θ = C / N is the observed concordance. θ estimates the probability of Identity by Descent, that is, that the concordant alleles in the two brothers are copies of the same chromosomal allele in the mother. The Odds Ratio is then the probability of the observed combination of concordant and non-concordant markers relative to a random combination. Note that the null hypothesis C = D gives an odds ratio of unity and a LOD score of zero, and that LOD scores are symmetrical around this value. Linkage increases θ > 0.5, and linkage should affect several adjacent markers so long as they are close enough together. Deviations of θ < 0.5 can only be due to chance, and should not occurs in runs; the top of the table is truncated.
Estimation of θ is simple where there are exactly two alleles at each locus and the mother is heterozygous at every locus.The 33 : 7 ratio in the table above gives an exact θ = 0.825, similar to the adjusted 0.82 calculated for the same ratio from an actual X-chromosome study by Hamer et al. (1994). Calculation of θ is complicated by many genetic factors, in particular whether the mother's genotype is known or not and if so whether she is homozygous or heterozygous, the occurrence and frequencies of multiple alleles at the marker locus, sample size, etc.
For example, if the mother's genotype is available and is known to be heterozygous at every locus tested, the probability that any allele shared by two brothers is Identical by Descent is 0.5. If the mother is known to be homozygous, the brothers necessarily share an allele (Identical by State), and the locus is uninformative about linkage of one of the alleles to the trait. If the mother's genotype is unknown, and the two brothers share an allele, θ depends on the the probability that she carried that allele either as a heterozygote or homozygote, which in turn depends on the frequency of the allele in the population.
For a polymorphic locus with two alleles at unequal frequencies [e.g., q = 0.8 and p = (1 - q) = 0.2 ], the probability that she is heterozygous for the more common allele is (2)(1-q)(q) = (2)(0.2)(0.8) = 0.32 ] and that she is homozygous for the same allele is 0.82 = 0.64. Thus, if the mother's genotype is unknown and the two brothers share the more common allele, the ratio is 0.32 / 0.64 and the probability is only 33% that she was a heterozygote and the two alleles in her sons are Identical by Descent. A substantial adjustment must be made for the 67% probability that she was homozygous and the brothers have received alternate, non-concordant alleles. The probability that the mother was heterozygous for the less common allele (q = 0.2) is given by the ratio 0.32 / 0.04 and the probability is 87.5%: a smaller adjustment is required.
For a polymorphic locus with multiple alleles at small frequencies [e.g., 10 alleles at q = 0.1 each], the probability that the mother was heterozygous for any allele pair is (1 - (10)(0.12) = 0.9, and the probability that she is heterozygous including any particular allele is (2)(1-q)(q) = (2)(0.9)(0.1) = 0.18, which is substantially greater than the probability that she is homozygous for the same allele, 0.12 = 0.01. So, if the two brothers share one of these rare alleles and the mother's genotype is unknown, the ratio is 0.18 / 0.01 and the probability 95% that she was a heterozygote at this locus. Only a slight adjustment must be made for the possibility that she was homozygous.
The conventional approach to a single-classification problem (concordance or non-concordance, with either outcome of equal probability) would be a Chi-square test. Significant concordance in the above table occurs at 26:14, for which X2 = 3.60 and p ~ 0.05. However, where multiple locus comparisons are done simultaneously, about one concordance test in 20 is expected to be show a significant score, simply by chance. Further, the Chi-square test is a parametric test that assumes a certain distribution of linkage relationships, which may not be so. Thus the preferred statistically approach is an odds ratio.
For each marker, the Odds Ratio = [ (θC)(1-θ)D ] / 0.5C+D , where C is the number of concordant pairs, D the number of discordant pairs (thus N = C+D), and θ = C / N is the observed concordance. θ estimates the probability of Identity by Descent, that is, that the concordant alleles in the two brothers are copies of the same chromosomal allele in the mother. The Odds Ratio is then the probability of the observed combination of concordant and non-concordant markers relative to a random combination. Note that the null hypothesis C = D gives an odds ratio of unity and a LOD score of zero, and that LOD scores are symmetrical around this value. Linkage increases θ > 0.5, and linkage should affect several adjacent markers so long as they are close enough together. Deviations of θ < 0.5 can only be due to chance, and should not occurs in runs; the top of the table is truncated.
Estimation of θ is simple where there are exactly two alleles at each locus and the mother is heterozygous at every locus.The 33 : 7 ratio in the table above gives an exact θ = 0.825, similar to the adjusted 0.82 calculated for the same ratio from an actual X-chromosome study by Hamer et al. (1994). Calculation of θ is complicated by many genetic factors, in particular whether the mother's genotype is known or not and if so whether she is homozygous or heterozygous, the occurrence and frequencies of multiple alleles at the marker locus, sample size, etc.
For example, if the mother's genotype is available and is known to be heterozygous at every locus tested, the probability that any allele shared by two brothers is Identical by Descent is 0.5. If the mother is known to be homozygous, the brothers necessarily share an allele (Identical by State), and the locus is uninformative about linkage of one of the alleles to the trait. If the mother's genotype is unknown, and the two brothers share an allele, θ depends on the the probability that she carried that allele either as a heterozygote or homozygote, which in turn depends on the frequency of the allele in the population.
For a polymorphic locus with two alleles at unequal frequencies [e.g., q = 0.8 and p = (1 - q) = 0.2 ], the probability that she is heterozygous for the more common allele is (2)(1-q)(q) = (2)(0.2)(0.8) = 0.32 ] and that she is homozygous for the same allele is 0.82 = 0.64. Thus, if the mother's genotype is unknown and the two brothers share the more common allele, the ratio is 0.32 / 0.64 and the probability is only 33% that she was a heterozygote and the two alleles in her sons are Identical by Descent. A substantial adjustment must be made for the 67% probability that she was homozygous and the brothers have received alternate, non-concordant alleles. The probability that the mother was heterozygous for the less common allele (q = 0.2) is given by the ratio 0.32 / 0.04 and the probability is 87.5%: a smaller adjustment is required.
For a polymorphic locus with multiple alleles at small frequencies [e.g., 10 alleles at q = 0.1 each], the probability that the mother was heterozygous for any allele pair is (1 - (10)(0.12) = 0.9, and the probability that she is heterozygous including any particular allele is (2)(1-q)(q) = (2)(0.9)(0.1) = 0.18, which is substantially greater than the probability that she is homozygous for the same allele, 0.12 = 0.01. So, if the two brothers share one of these rare alleles and the mother's genotype is unknown, the ratio is 0.18 / 0.01 and the probability 95% that she was a heterozygote at this locus. Only a slight adjustment must be made for the possibility that she was homozygous.