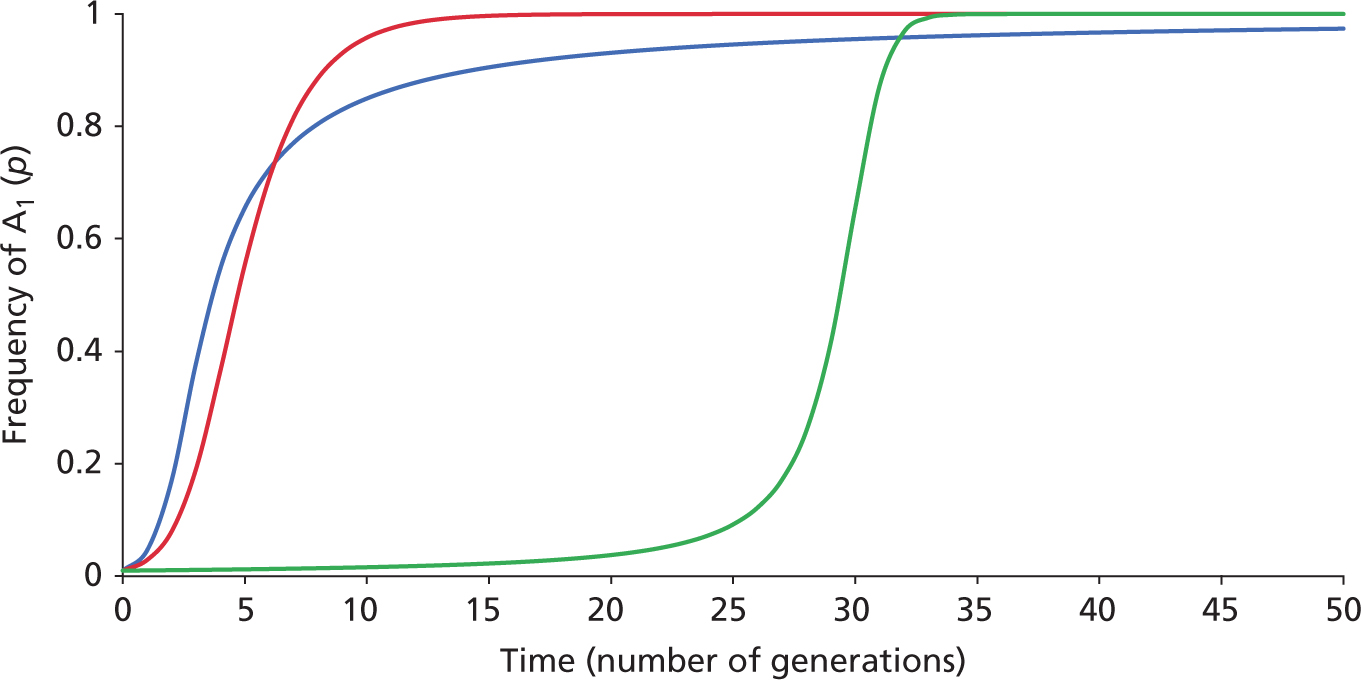
Change in frequency
of a rare allele under Positive
Directional Selection
Dominant, Semi-Dominant, & Recessive cases
In a single-locus model
with two alleles A1 and A2,
initial p = f(A1)
= 0.001. The three curves
trace f(A1)
over time for three
modes of dominance relationships.
The Blue curve
shows the case of dominance of
A1
to A2,
such that W11
= W12 = 1.0
and W22 = 0.2.
The Red curve
shows the additive (semi-dominance) model, in
which each A2 allele
decreases dW = 0.4, such that W11 = 1.0,
W12
= (1.0 - 0.4) = 0.6, and W22
= 1.0 - (0.4 + 0.4) = 0.2. The Green curve shows
the case where
A1
is recessive to
A2
, such that W11
= 1.0 and W12
= W22 = 0.2.
The difference between the shapes of the curves reflects
how mean population fitness
(![]() )
varies as f(A1)
)
varies as f(A1) ![]() 1.0
(SR2019 4.2).
1.0
(SR2019 4.2).
The dominance relationships
of any two alleles at a locus are fixed genetically.
The graph can also illustrate the fate of a common allele
under negative directional selection: invert the
Y-axis values top to bottom (1 ![]() 0)
and label it f(A2) = q.
That is, the mathematical behaviors of advantageous and
disadvantageous alleles are complementary for
any particular dominance model.
0)
and label it f(A2) = q.
That is, the mathematical behaviors of advantageous and
disadvantageous alleles are complementary for
any particular dominance model.
The principles presented in
this graph will be explored in greater depth in the
laboratory exercises for Natural Selection.
HOMEWORK:
Demonstrate that these same curves can be obtained for f(B)
= q, from appropriate values entered in the Hardy-
Weinberg program GSM in Excel.