Basic Concepts of Coalescence: Complete Coalescence
Coalescence
is the idea that all genetic variation in a
finite population is lost over finite time. Looked at backwards
in time, a population of N individuals after
t generations will all be descended from a single
individual in the earliest generation, and thus will have
only the allele(s) present in that individual, subject to
mutation.
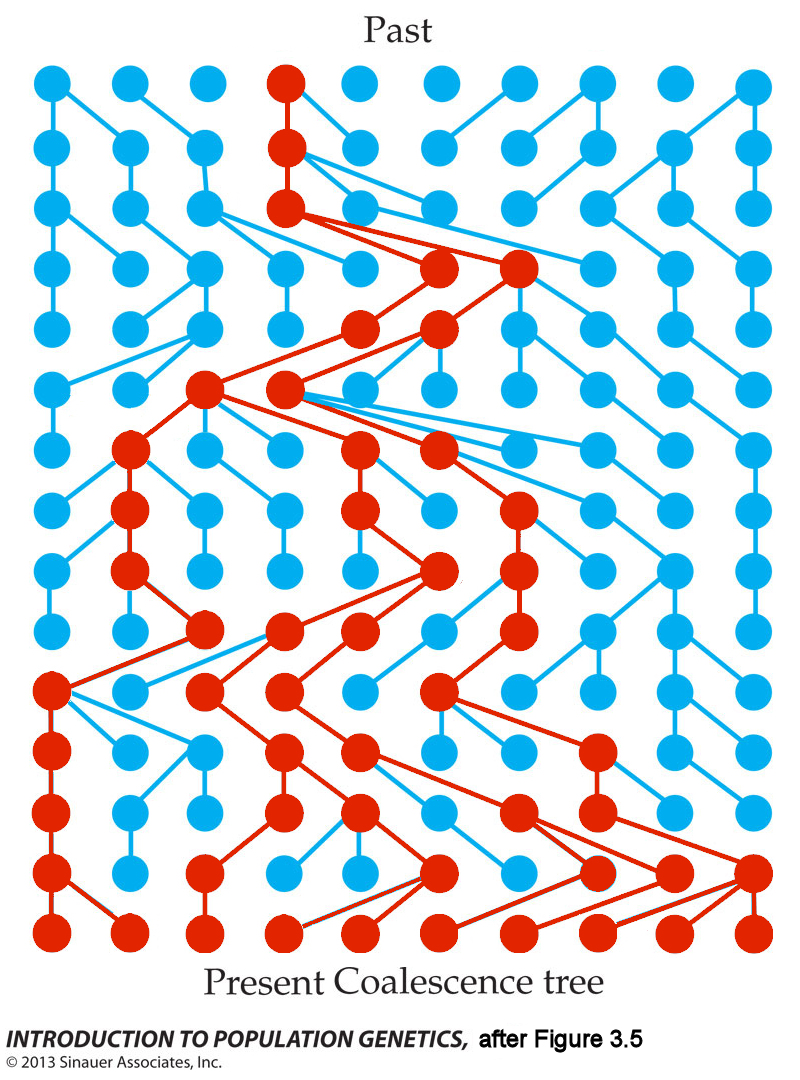
The array shows a population of haploid individuals that has a constant size of N=10 for 15 generations. Each horizontal row represents a generation; time proceeds from top to bottom. Lines in the graph show parent-to-offspring descent. All individuals in the Present (bottom row) are shown in red: if they are traced backwards in time (upward), all ten coalesce in a single Most Recent Common Ancestor (MRCA) in the third generation from the top. That is, all other variant alleles present in the third generation have been eliminated in the 15th generation.
Coalescence does not require change in population size, which remains constant at N=10 over the entire interval. Nor does it require a selective advantage for the coalescent ancestor. Ordinary (Poisson) variance in expected offspring number is sufficient, where approximately 1/3 of individuals will produce 0, 1, or 2 or more offspring in the next generation.
HOMEWORK: (1) For each of the other nine individuals in the first generation, identify the individual at which the lineage disappears.
(2) Some descendants of the MRCA in generation 3 are not shown in red: how do you interpret this?
(3) Redraw the descendants of the MRCA in lineages that lead to the present: how many separate lineages are present in each generation?
(4) Identify the error(s) in NS 03-02.
The array shows a population of haploid individuals that has a constant size of N=10 for 15 generations. Each horizontal row represents a generation; time proceeds from top to bottom. Lines in the graph show parent-to-offspring descent. All individuals in the Present (bottom row) are shown in red: if they are traced backwards in time (upward), all ten coalesce in a single Most Recent Common Ancestor (MRCA) in the third generation from the top. That is, all other variant alleles present in the third generation have been eliminated in the 15th generation.
Coalescence does not require change in population size, which remains constant at N=10 over the entire interval. Nor does it require a selective advantage for the coalescent ancestor. Ordinary (Poisson) variance in expected offspring number is sufficient, where approximately 1/3 of individuals will produce 0, 1, or 2 or more offspring in the next generation.
HOMEWORK: (1) For each of the other nine individuals in the first generation, identify the individual at which the lineage disappears.
(2) Some descendants of the MRCA in generation 3 are not shown in red: how do you interpret this?
(3) Redraw the descendants of the MRCA in lineages that lead to the present: how many separate lineages are present in each generation?
(4) Identify the error(s) in NS 03-02.
Figure after © 2013 by Sinauer; Text material © 2022 by Steven M. Carr