DNA Re-Sequencing Microarrays
(SM Carr et
al. 2008. Comp.
Biochem Physiol. D, Genomics & Proteomics 3,1-11)
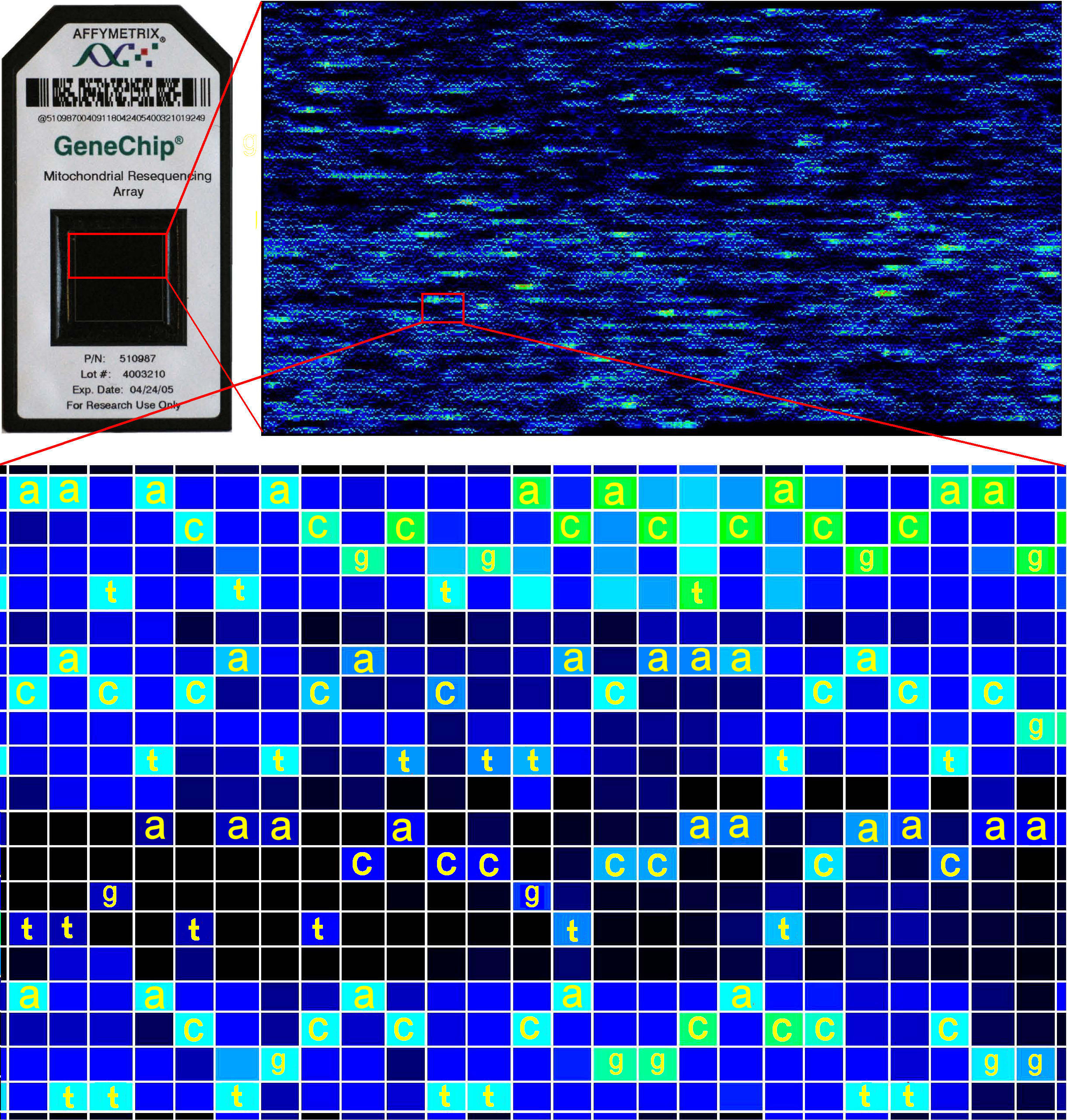
A DNA Re-sequencing microarray
allows a contiguous sequence of 30
~ 300Kbp to be determined in a single experiment. The microrarray is designed from a known
reference sequence, and allow the homologous sequence to be read
repeatedly from additional
individuals ("re-sequenced").
Re-sequencing is thus particularly well-suited to population genomics.
The microarray represents a reference sequence
of length n bases as a
series of 4 x n overlapping
("tiled") oligonucleotide
probes ("oligos"). For each 25-base oligo , three variant
probes are included that vary the middle base, one for each of the
three alternative DNA code
letters. Miss-match at
this position will prevent binding, so that a genomic DNA fragment with a SNP
sequence will stick to only one of the four oligos at any tiled position. [Click here for
further details]. The example shows a 15,452bp human mtDNA sequence in
a 320 row x
488 column array that includes both the sense and
antisense strands. Each nucleotide
position is represented in a vertical block of 4 cells in 5 rows (ACGT + a blank). In each
block, the cell with the
strongest relative intensity of DNA
binding identifies the base present at that position. In magnified
view, the sequence of bases in each of four blocks is easily read as the
left-to-right order of successive brightest 'spots'.
Where homologous mtDNA sequences differ by
> 20%, it is possible to include reference DNA sequences from several
species on the same microarray, and determine the sequences of
experimental DNA from
multiple individuals simultaneously. The ArkChip
is a high-throughput, cost-effective platform for conducting
several commercial and conservation projects simultaneously.

