
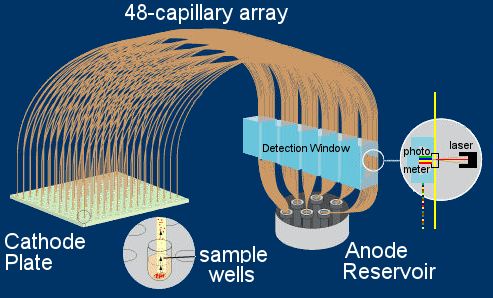
Automated DNA sequencing by capillary electrophoresis
Separation
of the products of a radioactively-labelled
or fluorescent dideoxy
sequencing reaction has historically been
done in a poly-acrylamide
slab gel, for example the kind used in a 96-lane ABI
377. Drawbacks off this method include the
necessity of preparing a fresh gel for each sequencing run,
manual loading of samples into the gel, relatively long run
times (8 ~ 10 hrs for resolution of a 1Kb DNA fragment), and
difficulties in automated and/or manual tracking of lanes.
An alternative
separation method uses capillary electrophoresis.
A series of standard dideoxy DNA
sequencing reactions are prepared, each in one well of an
8 row x 12 column 96-well
plate. [Left, A] A series of ultra-thin
capillary tubes (0.1 mm inside bore, 50 ~ 80 cm length)
are filled with a resin bead mixture and placed in a multi-capillary
array. One end of the array is mounted in a
negatively-charged Cathode
plate [right] so that their ends align with each well
of the sample plate. Application of high voltage causes the
labelled DNA in each sample
well to enter the corresponding capillary, and migrate
though it toward the positively-charged Anode reservoir. Because of
the high voltages used, electrophoretic separation requires only
1 ~ 3 hrs. As in slab-gel electrophoresis, the smaller fragments
move more quickly than the larger [Left, B]. The end-label dyes are
activated by a laser in the detector window
[right], and the fluorescence wavelength of each fragment is
read by a photometer as it passes a fixed point. Unlike
the slab gel system, the laser and photometer are fixed and
immovable, and can activate and read multiple side-by-side
capillaries simultaneously as separate data streams: this avoids
the problem of tracking separate lanes in a slab gel. Mobility
data are sent to a computer, and a chromatogram is
generated for each sample that is essentially identical to those
of the slab gel system.
Capillary sequencers typically use a robot to load a stack of sample plates automatically. The capillary array is flushed with buffer between plates, so that the machine can be left to run unattended for many runs. In the largest facilities, PCR reactions may also be linked directly to sequencing plates in robotic workstations that service dozens or hundreds of capillary sequencers. The chromatogram data are placed on a server, and can be downloaded by users hundreds or thousands of miles away. Economices of scale can make it cheaper to outsource DNA sequencing rather than buying, maintaining, and staffing an in-house machine.
All text material ©2015 by Steven M. Carr