transitions - alternative pyrimidines [ C
transversions - purine
Most mutations are transitions: interchanges of bases of same shape
 heritable meiotically between generations
heritable meiotically between generations
Nucleotide
interchanges are of two types
transitions - alternative
pyrimidines [ C![]() T ] or purines [ A
T ] or purines [ A![]() G ]
G ]
transversions - purine ![]() pyrimidine [C / T
pyrimidine [C / T
![]() A /
G]
A /
G]
Most mutations are transitions: interchanges
of bases of same shape
Spontaneous mutation
tautomeric shift - spontaneous,
transient rearrangement to alternative form
[IG1 16.07]
keto (standard) ![]() enol (rare) forms of
G & T
enol (rare) forms of
G & T
amino (standard) ![]() imino (rare) forms of A & C
imino (rare) forms of A & C
Non-standard bases have
altered pairing rules :
modified purine pairs with "wrong" pyrimidine
modified pyrimidine pairs with "wrong" purine
T' (enol)
pairs with G (keto)
C' (imino)
pairs
with A (amino)
G' (enol)
pairs with T (keto)
A' (imino)
pairs
with C (amino)
A tautomeric shift produces a transition mutation
in the complementary strand [see example]
[cf IG1 16.12]:
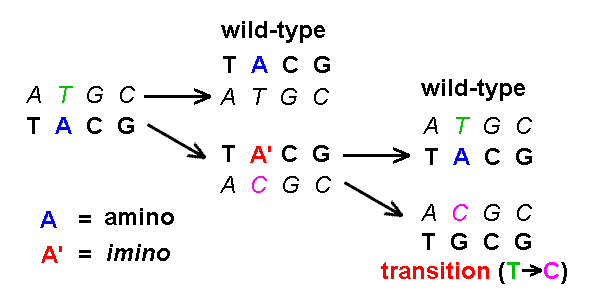
:
Based on this diagram, show that tautomeric shifts of the C,
G, & T bases also produce transition
mutations
Induced mutation - chemical modification of DNA alters base pairing [IG1 16.11, Table16.01]
alkylation - addition of alkyl group (methyl
CH3- or ethyl CH3-CH2-)
ethyl methane sulfonate
(EMS) is a common
laboratory mutagen
G ![]() 6-ethyl G
, pairs with T
6-ethyl G
, pairs with T
deamination - conversion of amino ![]() keto group [IG1 16.13]
keto group [IG1 16.13]
nitrous acid (HNO2)
is a common food additive / preservative
C ![]() U by
loss of NH2, pairs with A
U by
loss of NH2, pairs with A
A![]() hypoxanthine
by loss of NH2, pairs with C
hypoxanthine
by loss of NH2, pairs with C
depurination - loss of purine (A or G)
base in intact nucleotide
produces an apurinic
site
a common form of damage in 'ancient' DNA
[IG1 16.16]
replacement
of
base
may
produce
transversion
apyrimidinic sites are similar: loss
of C or T
intercalation - base analogue
"wedged" into DNA helix
[IG1 16.14]
acridine dyes resemble 3-ring base pair
intercalated analogue read as 'extra'
base:
DNAPol "stutters"  frameshift mutations
frameshift mutations
ethidium
bromide is a fluorescent intercalating dye of DNA
ionizing radiation: radiochemical damage to
DNA
Chromosome
damage
double - strand breaks :
non-homologue "sticky ends"
join end-to-end to form dicentric
chromosomes
226Ra - radium ingestion by watch dial painters
cross - linking - different DNA
molecules covalently joined
H-bonds converted to covalent bonds
Huntington Disease (OMIM citation 143100)
Neuro-muscular degeneration, 'choreic' movements,
typically with late
onset.
Huntingtin [sic] (HTT) locus has (CAG)n
repeat near 5'
end [4P16.3]
# copies in affected individuals inversely
correlated with severity & age of onset
9 ~ 36 unaffected,
36
~
41 incomplete penetrance
>41
adult onset; juvenile onset with >50
huntingtin protein with (gln)n
repeat has been isolated
[IG1 16.10]
autosomal dominant inheritance:
phenotype
is
a
consequence
of
presence
of modified protein:
 HD alleles 'dominate' standard alleles
HD alleles 'dominate' standard alleles
Fragile-X Syndrome [Martin-Bell
Syndrome] (OMIM
citation 300624)
Most common form of inherited
predisposition to mental retardation
(CGG)n repeat
occurs upstream from 5'
end of FMR-1 gene [Xq28]
6 ~
54 unaffected,
55 ~ 200 X-linked
female carriers, unaffected
>
200 affected sons