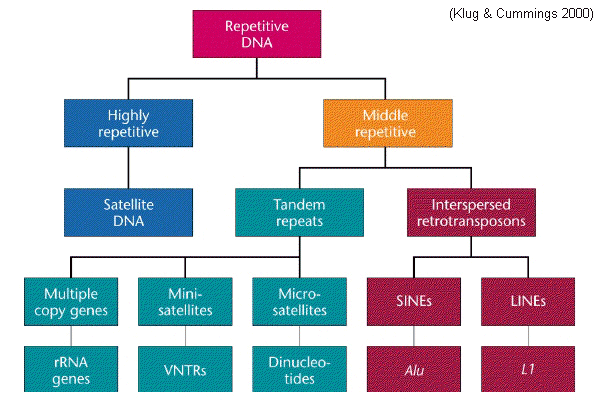
Categories of repetitive DNA in the human genome
More than half of the
human genome
consists of non-protein-coding,
repetitive
DNA elements. These occur in several classes
with
specific characteristics. The most important of these
are highly-repetitive
DNAs that
are detected as satellite DNA
in ultracentrifuge
experiments.These may be arranged either as
tandem
repeats
where specific short DNA
sequences
are repeated in end-to-end arrays from a few to many
tens of times, or as interspersed repeats
in which a particular sequence occurs at hundreds or
thousands of separate locations. Tandem microsatellites comprise
two-letter motifs, minisatellites
typically comprise four- or six-letters
motifs; both occur in variable numbers per locus.
Interspersed repeats may be short (SINEs) or long (LINEs),
for which the characteristic human families are Alu
(200~300 bp) and L1 (6,400 bp),
respectively. Other important SINEs include the
Alphoid (170 bp) family that constitutes
centromeric heterochromatin.