Analytical Ultra-centrifugation
Most
genetics textbooks illustrate the Meselson - Stahl experiment as though it were
conducted in a conventional preparative centrifuge. Preparative
centrifugation
applies low to moderate centrifugal force
(typically < 50,000 x g) to separate the heavier
components of a solution as a solid sediment at the bottom of the tube and a
liquid supernatant above,
either or both of which may be analyzed according to the
experiment. Analytical
centrifugation, the method used by Meselson &
Stahl, instead uses
extremely high centrifugal force (typically >> 100,000 x g) to
separate high-molecular weight sub-cellular molecules or
organelles that differ only slightly in density.
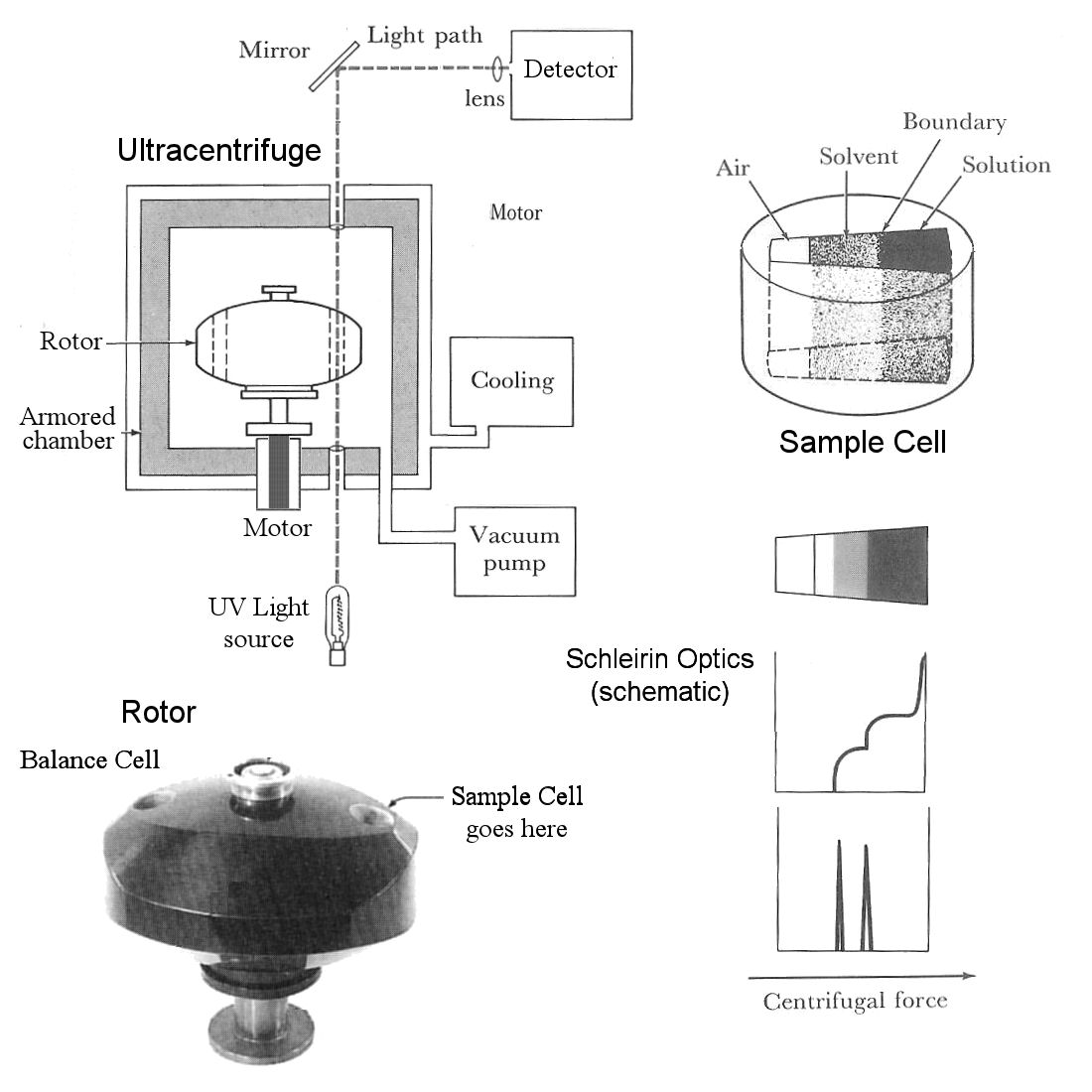
The sample to be analyzed is placed in a single sample cell (upper right), which is analogous to a centrifuge tube at a permanent right angle to the axis of rotation. The wedge shape means the lines of force at the walls of the cell are precisely parallel to the axes of centrifugal force. The sample cell is placed in a cylindrical hole on one side of the rotor, balanced by a blank cell with the same weight on the opposite side.
The rotor [bottom left] is placed on a spindle attached to a motor below the centrifuge. The rotor chamber is evacuated with a vacuum pump to reduce air friction, and cooled with a cooling jacket to maintain a constant temperature and prevent overheating. The chamber is surrounded with thick steel armor to contain the fragments in the event of a rotor failure. Progress of the experiment is monitored by passing a beam of light (UV in the case of nucleic acid experiments) through a quartz window in the bottom of the chamber, through transparent glass windows on the top and bottom of the sample cell, and out through another window in the top of the chamber, where it is directed by mirrors to a densitometer, camera, or other optical detection device. This is in effect a strobe light, except that the light is always on and a signal is detected only when the sample cell passes through the detection path.
The solution of molecules is introduced into the sample cell along with a solvent [top diagram]. The solvent in DNA experiments is typically cesium chloride (CsCl), which dissociates into high-density Cs+ ions that migrate outwards in the direction of the centrifugal force, forming a shallow density gradient. Molecules in solution diffuse centripetally [towards the axis] as the solution and solvent diffuse into each other. In the example [lower right], there are two molecular species, the lighter of which migrates faster and further [center-ward], and the denser of which lags behind. The discrete change in density at the interface between the two regions [middle diagram] bends the light passing through the sample cell at that point, and superimposition of adjacent signals (Schleirin Optical imaging) is seen as a dark spike at the transition point. The size and position of the spike are indications of quantity and molecular weight, respectively. [Imagine a Mexican Flag cocktail with grenadine, tequila, and creme de menthe layers].
Related methods are zonal analytical and preparative density gradient ultra-centrifugation.