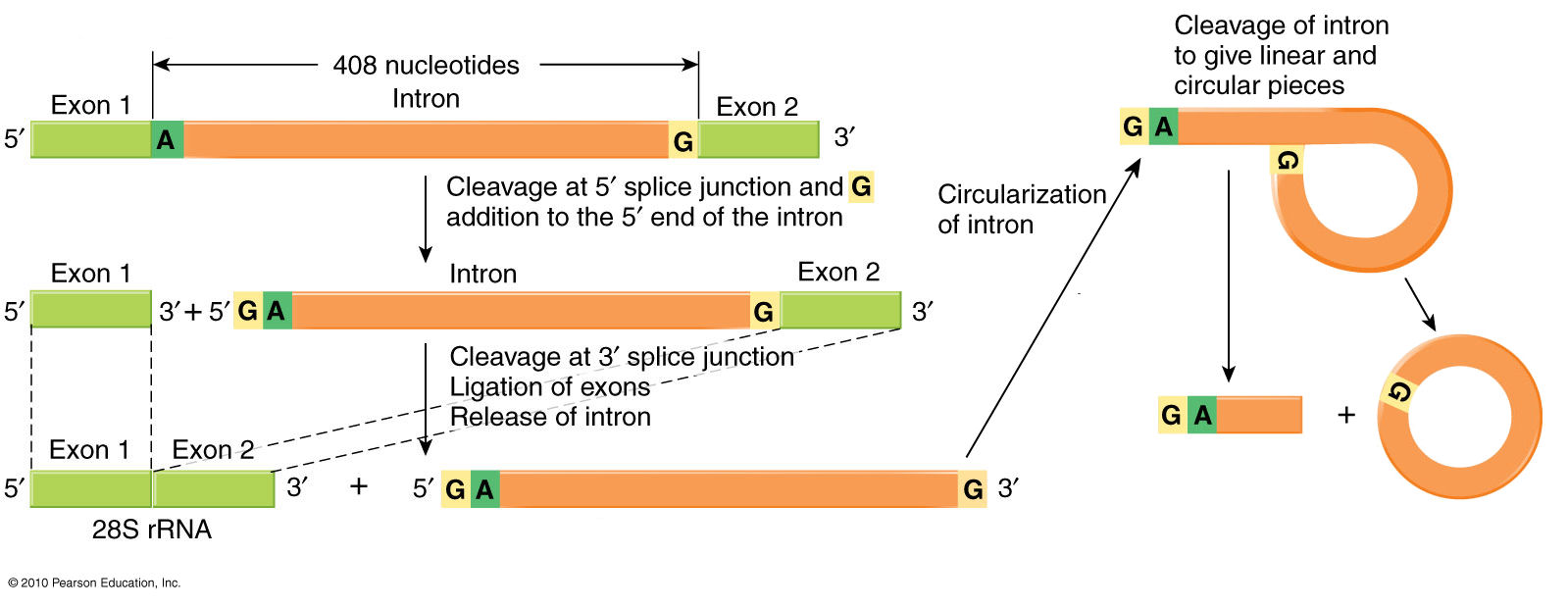
Exon
splicing
& intron excision in
eukaryotic hnRNA
The
5' & 3' ends of introns
have
characteristic GU and AG motifs that signal the region to
be "spliced
out*."
The diagram shows the removal of an intron between Exons 1 and
2. An
A
base near the AG signal
at
the 3' end of the
intron-equivalent
covalently bonds to the GU
signal at the junction of 5'
end of the intron and the 3'
end of Exon 1 [left
hand
diagram]. The 3'-OH
end of Exon 1 then
bonds to the 5'-PO4 of Exon
2 [right-hand diagram, below], which releases the
intron as a
covalently-bonded circular lariat with a
short tail. Note that the RNA
molecule
remains
unbroken throughout the process, and that it is the
splicing of the two exons that excises ('splices out') the
intervening
intron.
The sequences of the GU and AG intron motifs, and the flanking 3' & 5' bases of the exons, are critical to the process. Mutation of the corresponding bases in the DNA can result in mis-splicing, either failure to remove an intron or removal of an exon along with the adjacent intron.
[Notes on terminology: The dictionary definition of "splicing" is to join two things together at their ends, for example the ends of two ropes. It does not mean "splitting", as in separating two things. However, in molecular biology we speak of an intron transcript being "spliced out" of the final product.
The diagram refers to sequences in an hnRNA molecules as introns and exons. Properly speaking, introns and exons occur only in the DNA, and the corresponding RNA sequences are intron- and exon-equivalents].
The sequences of the GU and AG intron motifs, and the flanking 3' & 5' bases of the exons, are critical to the process. Mutation of the corresponding bases in the DNA can result in mis-splicing, either failure to remove an intron or removal of an exon along with the adjacent intron.
[Notes on terminology: The dictionary definition of "splicing" is to join two things together at their ends, for example the ends of two ropes. It does not mean "splitting", as in separating two things. However, in molecular biology we speak of an intron transcript being "spliced out" of the final product.
The diagram refers to sequences in an hnRNA molecules as introns and exons. Properly speaking, introns and exons occur only in the DNA, and the corresponding RNA sequences are intron- and exon-equivalents].