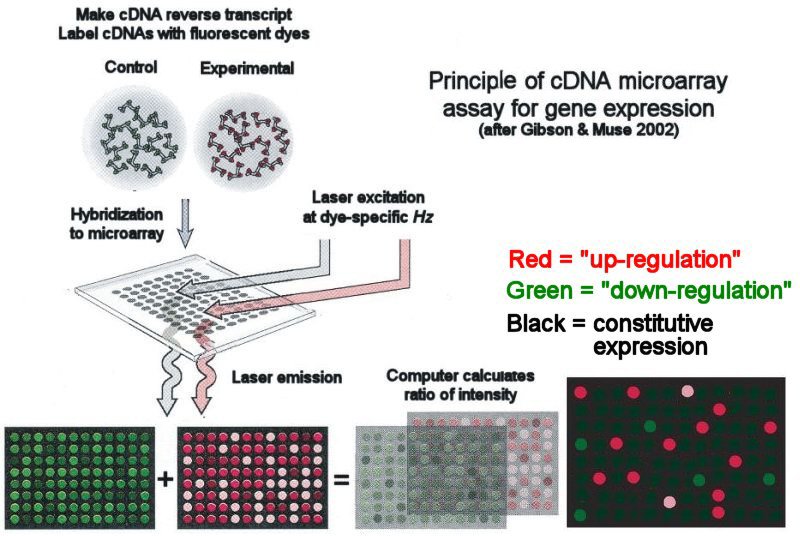

Principle of cDNA microarray assay of gene expression
A microarray is a set of short Expressed Sequence Tags (ESTs) made from a cDNA library of a set of known (or partially known) gene loci. The ESTs are spotted onto a cover-slip-sized glass plate, shown here as a 8x12 array. Microarrays with many thousand ESTs are now routine.
A complete set of mRNA
transcripts (the transcriptome)
is prepared from the tissue of an experimental
treatment or condition, e.g. fish fed a high-protein diet, or an
individual with breast cancer. Complementary DNA (cDNA) reverse
transcripts are prepared and labelled with a [red] fluorescent
dye. A control library
is constructed from an untreated source, e.g.
a standard fish
diet, or non-cancerous breast tissue; this library is labelled
with a different fluorescent [green]
dye. The experimental and control libraries are hybridized to
the microarray. A
Dual-Channel Laser
excites the corresponding dye, and the fluorescence intensity
indicates the degree of hybridization that has occurred.
Relative gene expression is measured as the ratio of the two
fluorescence wavelengths. Increased expression of
genes ("up-regulation")
in the experimental transcriptome relative to the control is
visualized as a "hot" red
"pseudo-color."
Decreased expression ("down-regulation")
shows as a "cool" green.
Intensity of either
color is proportional to the expression differential. Unchanged, constitutive expression (1:1 ratio of experimental
to control expression) shows as a neutral black.
In the linked theoretical example, cDNA microarrays are
used to measure life-stage
and
tissue specific patterns of gene expression.