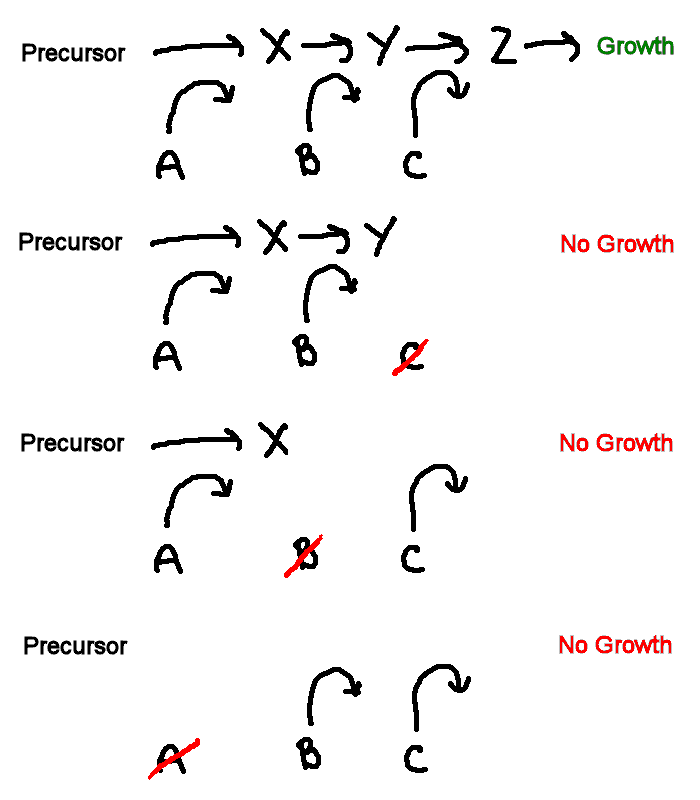
Inference of a biochemical pathway

This sketch shows a general model of a biochemical pathway. Substances X, Y and Z are intermediates in a metabolic pathway. Three enzymes A, B, & C convert some precursor to X, X to Y, and Y to Z, respectively. If all enzymes are present, the pathway is complete, and growth occurs (+). If any of the enzymes is missing or non-functional, growth cannot occur (-), irrespective of the presence of the other enzymes.
In the arginine
synthesis pathway
of Neurospora, the intermediate substances are the amino acids
arginine
(arg), citrulline (cit), and ornithine (orn).
Their relative order in the pathway is unknown. Given data showing the
growth response of a set of mutant strains to added amino acids, the
problem
is (1) to sort out the correspondence of
arg, cit,
and orn to X,
Y & Z, and (2) to
sort out the correspondence of the mutant strains
to defects in
enzymes A,
B, and C that interconvert these substances.
Beadle & Tatum
did their
experiments before the chemical nature of genes and gene mutations was
understood. Their insight was to connect the absence of a particular
enzymatic function with a defect in a particular "gene": "One Gene, One
Enzyme." We now
understand the molecular process by which genes direct the synthesis of
enzymes
(DNA makes RNA
makes Protein), and that enzyme defects
are due to alterations
of the DNA sequence (mutations).
It is
important to recognize that enzyme
defects, and the altered
phenotypes they produce, are consequences
of DNA mutations, not mutations
per se. Proteins do not mutate!