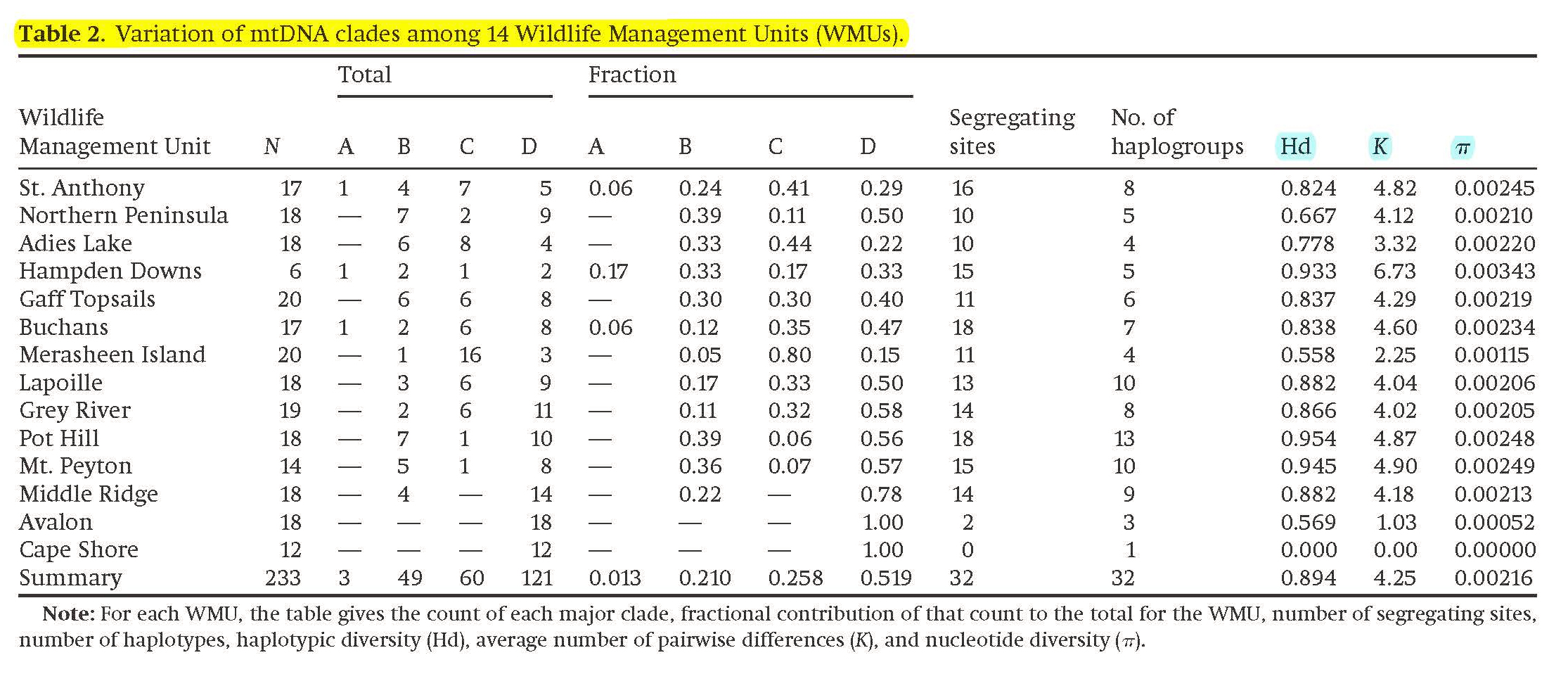
MtDNA sequence variation in Newfoundland Caribou by clade
Phylogenetic analysis of haplotypes show that they occur in four genetic lineages (clades), A, B, C, & D (Figure 1). The table gives counts and frequencies of each clade in each of 14 WMUs, and for each WMU the number of mtDNA sequences (haplogroups) as defined by the number of variable (segregating) SNP (Single Nucleotide Polymorphisms) sites.
Indices of variation include (1) Haplotypic Diversity (Hd), the probability that any two randomly chosen Caribou will have identical mtDNA sequences, (2) average pairwise difference (K), counted as the number of SNP differences for all (N)(N-1)/2 comparisons in the WMU, and (3) Nucleotide Diversity (π), which is K corrected for the number of nucleotides compared in each case.
mtDNA sequences are haploid: the data are similar to those for diploid nuclear alleles, neglecting the occurrence of the latter in genotypic pairs. For example, Hd and He can both be simulated by the 'random-draw-&-replacement model', except for a small correction in non-selfing diploids that two alleles may not be drawn from the same individual.
Table 2 from Wilkerson et al. 2018; Text material © 2025 by Steven M. Carr