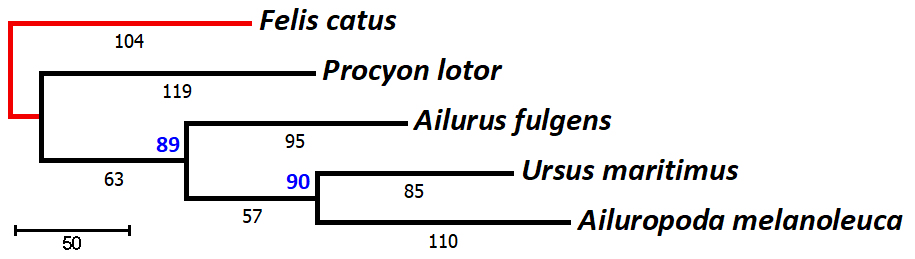
Outgroup Rooting
of an unrooted network

Outgroup Rooting
of an unrooted network
Given an unrooted
network of relationships [based on the 1,140 bp DNA sequence
of mtDNA Cytochrome b gene] among four species
of Carnivora in the suborder Caniformia, outgroup rooting uses an
additional taxon (an outgroup)
known from independent evidence to be less closely related to
any of the other species (the ingroup)
than they are to each other. The analysis is repeated with the
five taxa, and the placement of the outgroup among the ingroup
determines the root.
In the example, Felis
is a carnivore in a different suborder (Feliformia)
than the other four species (Caniformia). Inclusion of Felis
in the network analysis places the root on the internode
between Procyon and Ailurus, as expected if Ursus
and Ailuropoda are each others closest relatives in
the bear family, Ursidae.
This method
requires accurate information as to ingroup / outgroup
relationships. The outgroup should be a sister
taxon, a closely related taxon known
not to be included in the ingroup. If a more distant
outgroup taxon is used, accumulated differences from the ingroup
may obscure the actual similarities. Note that in this example,
the bootstrap support for the branch leading to the two Ursidae
has fallen to 90% from
99% in the unrooted network.