Natural Selection on semi- & incompletely dominant phenotypes with Additive allele effects
In classical genetics, if the phenotype of the AB genotype
is intermediate between AA & BB, but
closer to that of the AA than the AB genotype,
A is described as incompletely
dominant to B. If the AB phenotype
is precisely intermediate between that of the two homozygous
genotypes, A and B are described as semi-dominant.
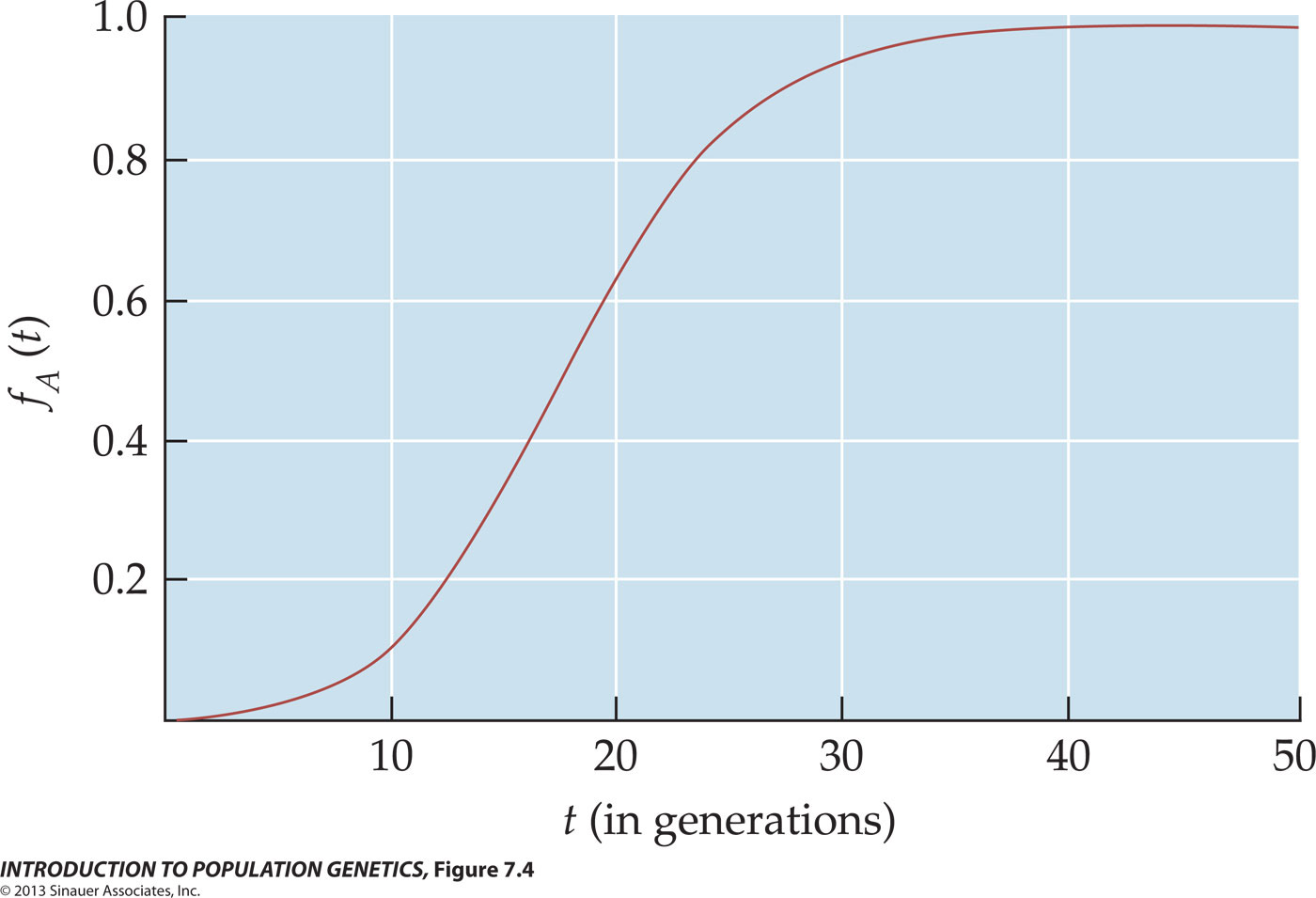
With respect to fitness phenotypes, semi-dominance occurs for example when genotypes AA, AB, & BB are assigned fitness values of W0 = 1.0, W2 = 0.8, and W2 = 0.6. With the notation of selection coefficients, the fitness values would be written as WAA = 1, WAa = (1 - s), and Waa = (1 - 2s), where s = 0.2. That is, each B allele contributes an additive selective disadvantage of s = 0.2, so that a BB homozygote is at twice the disadvantage of the AB heterozygote.
In the table below, let initial f(A) = 0.01. Note that if s > 0.5, fitness of AB heterozygotes W2 < 0 and therefore undefined. At s = 0.5, the alternative B allele is semi-lethal. Compare this model with that for multiplicative Genic Selection.
Note once again that A is semi-dominant to B, not because it has superior fitness (and might be said incorrectly to "dominate" the other allele), but because the AB phenotype is intermediate between that of the AA and BB. Genetic dominance is a genotypic, not a phenotypic, relationship.
Simple additive dominance may be typical at many gene loci, where the phenotype is a consequence of the equal contribution of both alleles. For example, the amount of gene product. Incomplete dominance may also be typical at other gene loci, where the phenotype is (much) more strongly influenced by one allele than the other. For example, given a null allele that produces no gene product, the standard allele may be "up-regulated" so that the amount of gene product in the AB heterozygote is identical or much closer to that of the AA homozygote. It remains a point of contention what fraction of allelic variation that contributes to heterozygosity has any measurable effect on the observed phenotype relative to the homozygote. The so-called "Neutralist - Selectionist" controversy will be discussed elsewhere in the course.
HOMEWORK:
(1) For an initial f(A) = 0.01 and s≼
0.5
(2) For an initial f(A) = 0.01 and s = 1/10 of the values below [i.e., shift the decimal in s one place to the right for s≼ 0.5
(3) How do you interpret the values obtained for s > 0.5.
With respect to fitness phenotypes, semi-dominance occurs for example when genotypes AA, AB, & BB are assigned fitness values of W0 = 1.0, W2 = 0.8, and W2 = 0.6. With the notation of selection coefficients, the fitness values would be written as WAA = 1, WAa = (1 - s), and Waa = (1 - 2s), where s = 0.2. That is, each B allele contributes an additive selective disadvantage of s = 0.2, so that a BB homozygote is at twice the disadvantage of the AB heterozygote.
In the table below, let initial f(A) = 0.01. Note that if s > 0.5, fitness of AB heterozygotes W2 < 0 and therefore undefined. At s = 0.5, the alternative B allele is semi-lethal. Compare this model with that for multiplicative Genic Selection.
Note once again that A is semi-dominant to B, not because it has superior fitness (and might be said incorrectly to "dominate" the other allele), but because the AB phenotype is intermediate between that of the AA and BB. Genetic dominance is a genotypic, not a phenotypic, relationship.
Simple additive dominance may be typical at many gene loci, where the phenotype is a consequence of the equal contribution of both alleles. For example, the amount of gene product. Incomplete dominance may also be typical at other gene loci, where the phenotype is (much) more strongly influenced by one allele than the other. For example, given a null allele that produces no gene product, the standard allele may be "up-regulated" so that the amount of gene product in the AB heterozygote is identical or much closer to that of the AA homozygote. It remains a point of contention what fraction of allelic variation that contributes to heterozygosity has any measurable effect on the observed phenotype relative to the homozygote. The so-called "Neutralist - Selectionist" controversy will be discussed elsewhere in the course.
HOMEWORK:
(1) For an initial f(A) = 0.01 and s
(2) For an initial f(A) = 0.01 and s = 1/10 of the values below [i.e., shift the decimal in s one place to the right for s
(3) How do you interpret the values obtained for s > 0.5.
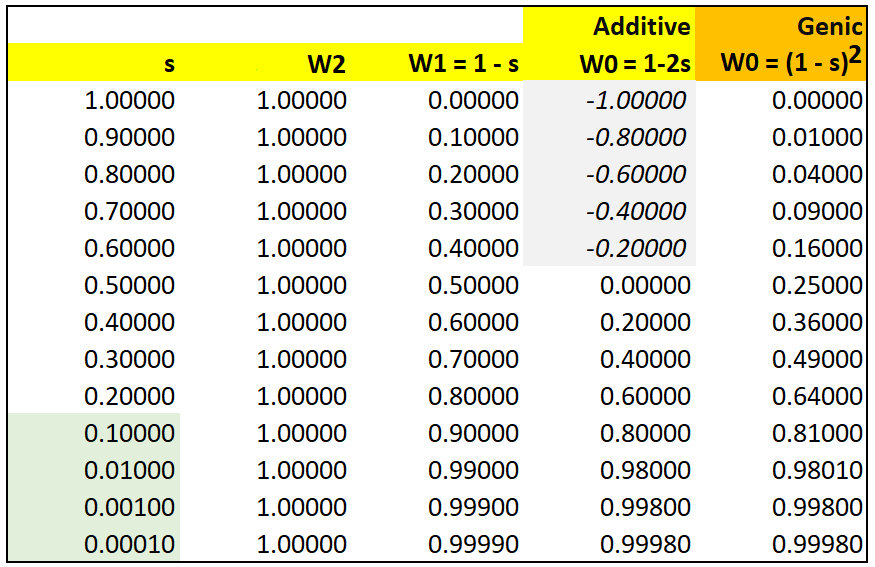
Figure © 2013 by Sinauer; Table & text material © 2025 by Steven M. Carr