Natural Selection on SNP mutation in Haploid Bacteria & Bacteriophages
Selection in a haploid system involves the relative
elective advantage of SNP haplotypes at a
locus, rather than those at a diploid genotype.
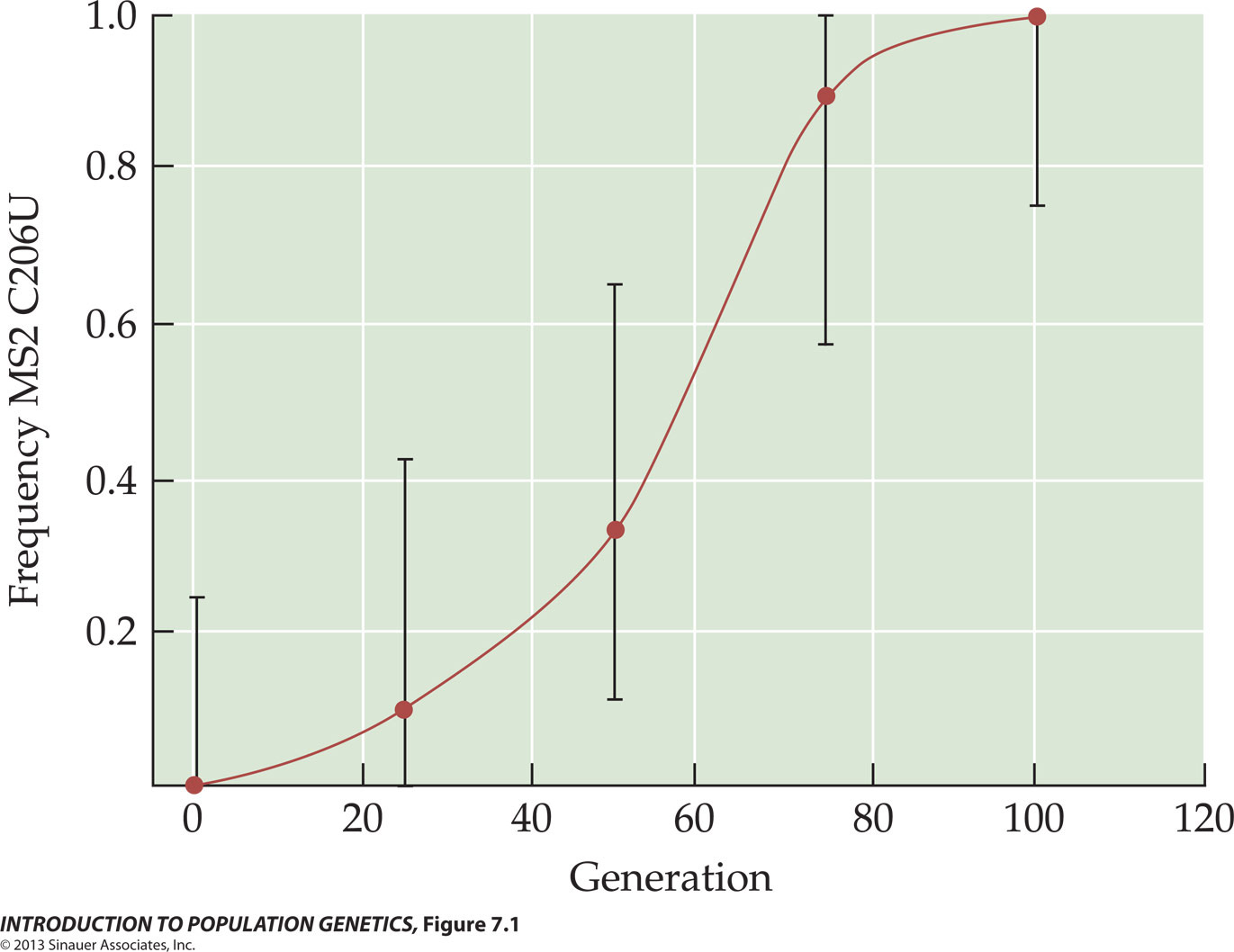
Here, in the MS2 RNA phage,
the newly arisen SNP mutation is C206U,
where the standard C base at position 206
has mutated to a U base. The U
haplotype has a selective advantage: the graph shows its
increase in vitro at intervals of 25 generations. The error bars show variation in the rate of
fixation among replicate cultures. The increase in
f(U) is exponential, and the
pre-existing C allele is the minority alleles in
some populations by 50 generations, eliminated in some
populations by 75 and in almost all by 100 generations.
Contrast this to the behavior of a disadvantageous allele in
a diploid system, where it is never entirely
eliminated because when rare it show up only in
heterozygotes.
The haploid selection model accounts for the increase of the 'Delta' & 'Omicron' variants of the Covid-19 RNA virus during the Covid-19 epidemic. In the data base, the original Alpha strain was replaced in vivo in the human species, first by the Delta and then by Omicron variants, because of their higher infectivity rate (Ro, read as "R nought").
The haploid selection model accounts for the increase of the 'Delta' & 'Omicron' variants of the Covid-19 RNA virus during the Covid-19 epidemic. In the data base, the original Alpha strain was replaced in vivo in the human species, first by the Delta and then by Omicron variants, because of their higher infectivity rate (Ro, read as "R nought").
Figure © 2013 by Sinauer; Text material © 2025 by Steven M. Carr