Calculation of Tajima's Estimator π
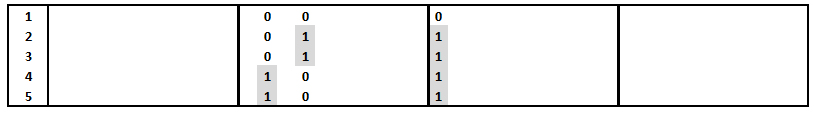
Recall the bottom panel from the sample DNA data
set, which shows three distinct binary coded
alleles (haplotypes) among five individuals: 000
in #1, 011 in ##2 & 3, and
101 in ## 4 & 5. Remember that 0
and 1 are coded differences, not numeric
values. Let us suppose these are the only variable
sites over four blocks of ten bases each (L
= 40).
We wish to estimate the mean number of pairwise differences among individuals within a population. This is Tajima's estimator π, defined as
The numerator is the count of all pairwise differences among sequences, the denominator is the number of pairwise comparisons. The latter is the combinatorial "n choose 2" = (n)(n-1)/2. With n = 5 sequences, there are (5)(4)/(2) = 10 pairwise comparisons. The numerator is then the sum of the number of differences for each of these 10 comparisons. The number of differences between Sequence #1 and ##2,3,4,5 = 2 + 2 + 2 + 2 = 8; between #2 and ##3,4,5 = 0 + 2 + 2 = 4; between #3 and ##4,5 = 2 + 2 = 4; between #4 and #5 = 0. Thus the total number of differences is 8 + 4 + 4 + 0 = 16. Then π = 16/10 = 1.6 pairwise differences.
The same result can be obtained by counting the number of pairwise difference between individuals at each variable position. In this example, there are 5, 6, & 4 differences at the three positions, for a total of 16, as above.
If we wanted to compare the average pairwise differences between two genes of different lengths, we could correct π for each gene by dividing by the length L of that gene in base pairs: in this case L = 40 and π' = 1.6 / 40 = 0.04 differences / bp.
Advanced Note: π is said to be an estimator of the value θ, which is the parametric expectation of the number of differences between two gene copies. Recall that a parameter is the actual value of something. The mathematics lurking behind this is the notion that if a gene comprised an infinite number of sites, then every new SNP mutation would create a new allele: this is called the Infinite Alleles Model. The virtue of the IAM is that it allows for simpler theoretical models, most of which were developed before actual DNA sequence data became available. The disadvantage of the model is that it is unrealistic, because all gene sequences are finite, there are a limited number of possible SNP sites, and in particular because SNP mutations may recur at any site. However, if differences among alleles are relatively small, the IAM is reasonably accurate.
We wish to estimate the mean number of pairwise differences among individuals within a population. This is Tajima's estimator π, defined as
π = dij
/ [(n)(n-1)/2]
The numerator is the count of all pairwise differences among sequences, the denominator is the number of pairwise comparisons. The latter is the combinatorial "n choose 2" = (n)(n-1)/2. With n = 5 sequences, there are (5)(4)/(2) = 10 pairwise comparisons. The numerator is then the sum of the number of differences for each of these 10 comparisons. The number of differences between Sequence #1 and ##2,3,4,5 = 2 + 2 + 2 + 2 = 8; between #2 and ##3,4,5 = 0 + 2 + 2 = 4; between #3 and ##4,5 = 2 + 2 = 4; between #4 and #5 = 0. Thus the total number of differences is 8 + 4 + 4 + 0 = 16. Then π = 16/10 = 1.6 pairwise differences.
The same result can be obtained by counting the number of pairwise difference between individuals at each variable position. In this example, there are 5, 6, & 4 differences at the three positions, for a total of 16, as above.
If we wanted to compare the average pairwise differences between two genes of different lengths, we could correct π for each gene by dividing by the length L of that gene in base pairs: in this case L = 40 and π' = 1.6 / 40 = 0.04 differences / bp.
Advanced Note: π is said to be an estimator of the value θ, which is the parametric expectation of the number of differences between two gene copies. Recall that a parameter is the actual value of something. The mathematics lurking behind this is the notion that if a gene comprised an infinite number of sites, then every new SNP mutation would create a new allele: this is called the Infinite Alleles Model. The virtue of the IAM is that it allows for simpler theoretical models, most of which were developed before actual DNA sequence data became available. The disadvantage of the model is that it is unrealistic, because all gene sequences are finite, there are a limited number of possible SNP sites, and in particular because SNP mutations may recur at any site. However, if differences among alleles are relatively small, the IAM is reasonably accurate.
Figure after © 2013 by Sinauer & Text material © 2022 by Steven M. Carr