Multi-species DNA re-sequencing chip: the "ArkChip"
(SM Carr et al. 2008. Comp. Biochem Physiol. D, Genomics & Proteomics 3,1-11)
The
first generation of re-sequencing chips accommodated 30,000 base
pairs (30Kbp) of reference DNA sequence,
enough for two 15Kbp mtDNA
genomes (without control regions). Since the 25b oligonucleotides are
species-specific,
re-sequencing of all included genomes can be accomplished
simultaneously. Separate
investigations on distinct species can be "multi-plexed."
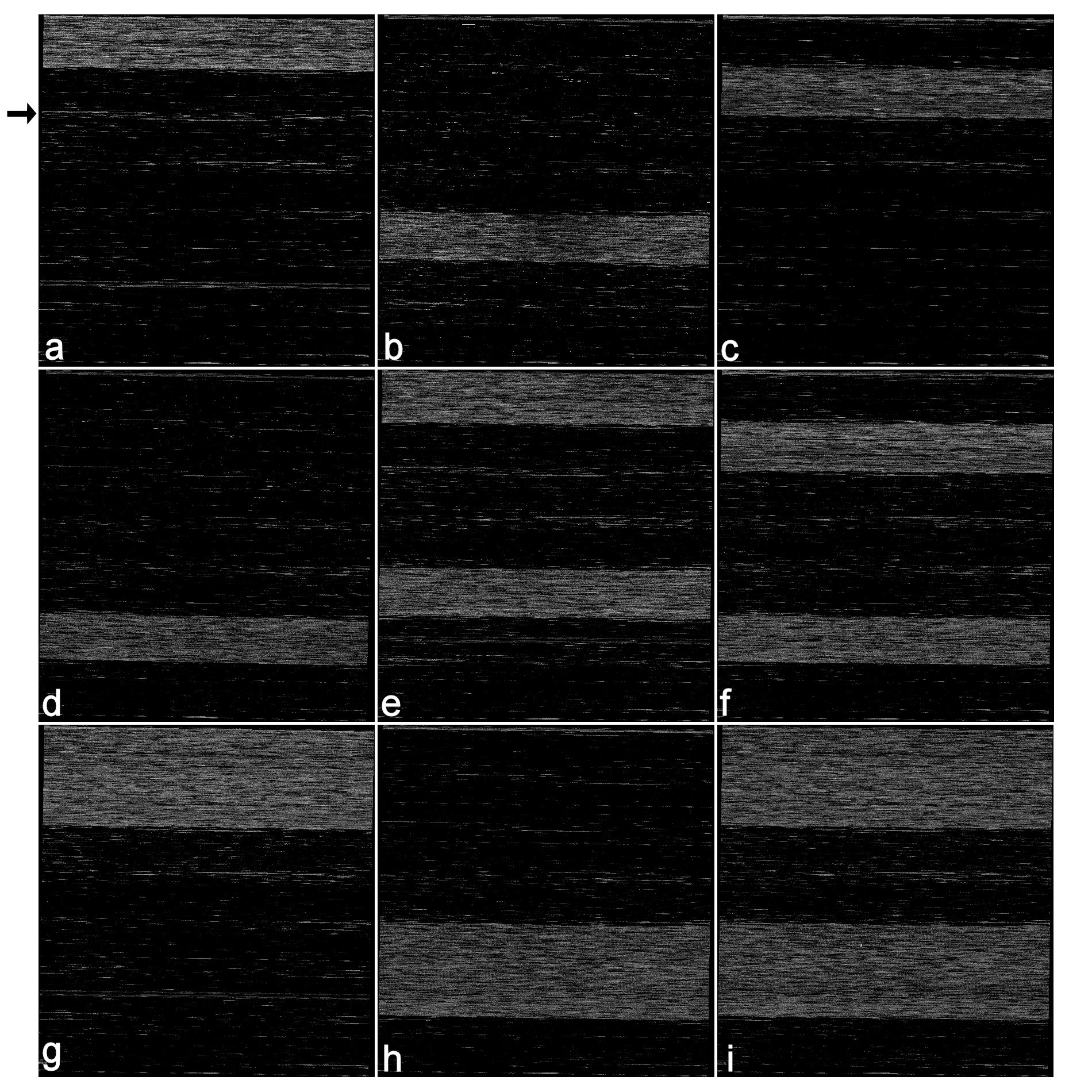
The figure shows the design of a multi-species iterative re-sequencing microarray—the “ArkChip”. The design for a 120-kbp chip includes the sense and antisense strands of the complete mtDNA genome sequence of three fish, three mammals, and one bird species. These are tiled on the array in seven successive blocks of oligonucleotides. The nine panels show the results of four separate single-species experiments with mtDNA from Atlantic Cod, Newfoundland Caribou, Atlantic Wolffish, and Harp Seals (blocks 1, 4, 2, & 6, respectively, in panels a-d, respectively), four pairwise experiments with cod + caribou, wolffish + seal, cod + wolffish, caribou + seal (panels e-h), and one with all four species (panel i). Note the species-specificity of mtDNA annealing in each experiment to the appropriate block(s). The arrow in panel (a) indicates a region of intermittent cross-hybridization to a conserved sequence tiled in another species, which occurs in other experiments as well.
By combining multiple species recovery projects in a single "ArkChip" design, initial design costs and per-experiment costs are reduced 16-fold: the cost of conducting a complete mtDNA genomic population analysis becomes comparable to that for a current single-locus project.
The figure shows the design of a multi-species iterative re-sequencing microarray—the “ArkChip”. The design for a 120-kbp chip includes the sense and antisense strands of the complete mtDNA genome sequence of three fish, three mammals, and one bird species. These are tiled on the array in seven successive blocks of oligonucleotides. The nine panels show the results of four separate single-species experiments with mtDNA from Atlantic Cod, Newfoundland Caribou, Atlantic Wolffish, and Harp Seals (blocks 1, 4, 2, & 6, respectively, in panels a-d, respectively), four pairwise experiments with cod + caribou, wolffish + seal, cod + wolffish, caribou + seal (panels e-h), and one with all four species (panel i). Note the species-specificity of mtDNA annealing in each experiment to the appropriate block(s). The arrow in panel (a) indicates a region of intermittent cross-hybridization to a conserved sequence tiled in another species, which occurs in other experiments as well.
By combining multiple species recovery projects in a single "ArkChip" design, initial design costs and per-experiment costs are reduced 16-fold: the cost of conducting a complete mtDNA genomic population analysis becomes comparable to that for a current single-locus project.
All material © 2008 by Steven M. Carr