DNA Sequencing by means of MALDI-TOF Mass Spectrometry
Mass Spectrometry
is an experimental technique used to identify the components of a
heterogeneous collection of biomolecules, by sensitive discrimination
of their molecular masses. In matrix-assisted
laser desorption / ionisation time-of-flight mass spectrometry (MALDI-TOF),
the sample to be analyzed is placed in a UV-absorbing matrix
pad and exposed to a short laser pulse. The ionized
molecules are accelerated off the matrix pad (desorption),
and move in an electric field towards a detector. The "time
of flight" required to reach the detector depends on the mass / charge (m / z)
ratio of the individual
molecules*.
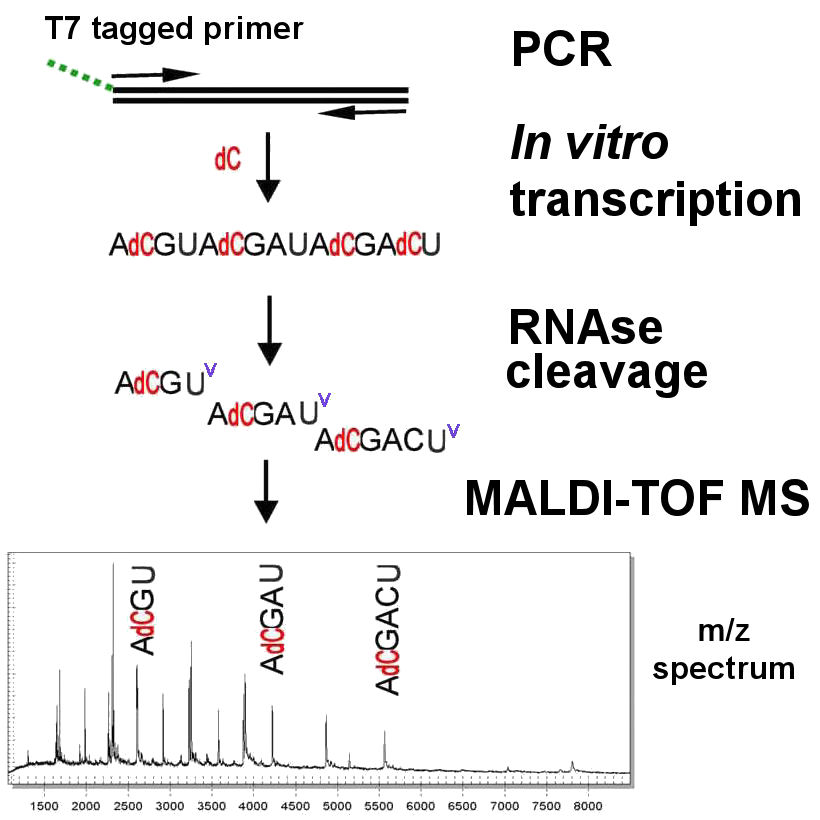
To use MALDI-TOF for DNA sequencing, consider an experiment in which the fragment ACGTACGATACGACT is to be sequenced. A PCR-derived DNA product is transcribed to RNA in vitro [ACGUACGAUACGACU] in four separate reactions, each with three rNTP bases and one specific dNTP. In the example, use of dC prevents cleavage of C positions by RNAse , which cleaves only after rU and produces three fragments of 4, 5, or 6 nucleotides. Each fragment has a characteristic m / z ratio, as indicated by a peak in the MALDI-TOF spectrum. Analogous reactions occur for each of the other three letters**. The MALDI-TOF mass signal pattern obtained for any experimental DNA sequence is compared with the expected m / z spectrum for the reference sequence under consideration, which includes the products of all four cleavage reactions. Any SNP differences between the experimental and reference DNA sequences will produce predictable shifts in the spectrum, and their exact nature can be deduced***.
*By analogy, the mass of a baseball thrown straight up can be deduced from the time in the air before it hits the ground, given knowledge of the force with which it is thrown.
** In greater detail: four transcription reactions are done with two forward and two reverse primers. In each pair, either a dC or a dT is used along with the other three rNTPs. Since RNAse cleaves only after rC and rU, incorporation of dC protects those bases and cleavage occurs only after rU. Use of dT allows cleavage only after rC. The same process on the complementary strand in the reverse reactions produces two fragment sets cleaved after rC and rU on the reverse strand, which corresponds to cleavage before rA and rG bases on the forward strand [think about it]. The four reactions taken together include a collection of fragments terminated adjacent to every base in the sequence, as in the example.
*** Generation of the expected spectrum of the reference sequence is an example of in silico genomics, that is, genomics aided by computer. Prediction of the effect of any given SNP difference on the spectrum is also done by computer.
To use MALDI-TOF for DNA sequencing, consider an experiment in which the fragment ACGTACGATACGACT is to be sequenced. A PCR-derived DNA product is transcribed to RNA in vitro [ACGUACGAUACGACU] in four separate reactions, each with three rNTP bases and one specific dNTP. In the example, use of dC prevents cleavage of C positions by RNAse , which cleaves only after rU and produces three fragments of 4, 5, or 6 nucleotides. Each fragment has a characteristic m / z ratio, as indicated by a peak in the MALDI-TOF spectrum. Analogous reactions occur for each of the other three letters**. The MALDI-TOF mass signal pattern obtained for any experimental DNA sequence is compared with the expected m / z spectrum for the reference sequence under consideration, which includes the products of all four cleavage reactions. Any SNP differences between the experimental and reference DNA sequences will produce predictable shifts in the spectrum, and their exact nature can be deduced***.
*By analogy, the mass of a baseball thrown straight up can be deduced from the time in the air before it hits the ground, given knowledge of the force with which it is thrown.
** In greater detail: four transcription reactions are done with two forward and two reverse primers. In each pair, either a dC or a dT is used along with the other three rNTPs. Since RNAse cleaves only after rC and rU, incorporation of dC protects those bases and cleavage occurs only after rU. Use of dT allows cleavage only after rC. The same process on the complementary strand in the reverse reactions produces two fragment sets cleaved after rC and rU on the reverse strand, which corresponds to cleavage before rA and rG bases on the forward strand [think about it]. The four reactions taken together include a collection of fragments terminated adjacent to every base in the sequence, as in the example.
*** Generation of the expected spectrum of the reference sequence is an example of in silico genomics, that is, genomics aided by computer. Prediction of the effect of any given SNP difference on the spectrum is also done by computer.
All material ©2008 by Steven M. Carr