S.
E. Luria and M. Delbruck (1943). Mutations of bacteria from vrius
sensitivity
to virus resistance. Genetics 28:491
Background
& Introduction
Max
Delbruck
(1906 -1981) & Salvador
Luria
(1912 - 1991) in 1941 & 1953
Shared 1969
Nobel Prize in Physiology or Medicine
Bacteriology
in 1940s not heavily
influenced by genetic thinking
Bacteria have no
nuclei:
do they have "genes"?
Bacterial "phenotypes"
are the manifestations of 106s of bacteria simultaneously
Bacteria don't have
sex: crosses not possible
[Discovery of bacterial sex led to 1958
Nobel Prize]
bacteriophages
("phages") - "subcellular living parasites that infect,
multiply
within, & kill bacteria."
T1
phages are active on E. coli
[phage] >> [bacteria]  no bacterial colonies grow: bacteria are Tons
("T-one sensitive")
no bacterial colonies grow: bacteria are Tons
("T-one sensitive")
[phage] ~ [bacteria]  some bacterial colonies grow: bacteria are Tonr
("T-one
resistant")
some bacterial colonies grow: bacteria are Tonr
("T-one
resistant")
Tonr phenotype
is stable
all descendant bacteria are Tonr
phenotype persists in the absence of T1
Two Hypotheses
1. Tonr phenotype
is induced by exposure of
bacteria
to phage
Each bacterium has finite chance of survival (say ~ 1 / 107);
Survivors have altered metabolic phenotype, which is
transmitted
to offspring
[distinction between phenotype
&
genotype
not clear]
Bacteria adapt to their environment
:
a Lamarckian hypothesis: inheritance of acquired
character
2. Tonr phenotype
occurs spontaneously, prior
to exposure of bacteria to phage
Some rare bacteria (say ~ 1 / 107) are already Tonr
These bacteria have undergone genetic mutation
to a stable genotype
[phenotype persists in
absence of phage]
a Darwinian hypothesis: Tonr bacteria
are
selected
Materials & Methods
Hypotheses make
different predictions as
to
numerical distribution
of Tonr phenotypes among bacterial cultures.
Induction
(Adaptation) Hypothesis
predicts: n / N = a
where n =
number
of Tonr bacteria observed out of
N = number of Tons bacteria plated, and
a = probability of conversion
from Tons to Tonr
Then, n should be
a constant fraction of N
Mutation
Hypothesis predicts: n
/ N = ga2g / 2g =
ga
where a = mutation rate
(# mutations / cell / generation)
g = # generations to go from 1  N
bacteria, so that
N
bacteria, so that
N = 2g doublings occur, of which
n = ga2g produce mutant Tonr
bacteria
[because a mutation in the i th generation contributes a2i2g-i
= a2g mutants]
Then, n should increase wrt N , as g
increases
How
can differences in n
be evaluated?
Suppose c
cultures
are started from a single Tons mutant each
after g
generations
there are N = 2g
bacteria in each culture
mean number of
Tonr
bacteria is 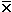 =
=  (xi)
/ c
(xi)
/ c
variance is 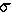 2
=
2
=  [
[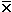 - xi]2 / c
- xi]2 / c
Thought
experiment:
Consider
four cultures started from a single bacterium
after g = 3 generations: expect 16 cells from 15 divisions @,
total 64 cells from 60 divisions
plate each culture separately w/ T1, count total # Tonr
Suppose 10
Tonr
colonies
observed
Induction
Hypothesis:
Tonr induction
occurred only in third generation by exposure to T1
probability of
induction
(a) is uniform / bacterium
a
= 10 inductions / 64 cells = 15%
mean rate of occurence = 10 / 4 = 2.5 Tonr
per
culture
Poisson
Distribution: variance = mean
variance = [(2.5 - 3)2
+ (2.5 - 1)2 + (2.5 -
5)2 + (2.5 - 1)2]
/ 4 = 2.19
Mutation
Hypothesis
Tonr mutation
has occurred spontaneouly, prior to exposure to T1
mutation rate
(a) = 2 events / 60 cell divisions = 0.033 mutations /
cell
/ generation
mean rate of occurence = (2 + 0 + 8 + 0) / 4 = 2.5 Tonr
as before
earlier Tonr mutations leave
more offspring (as in Culture
3)
variance
= [(2.5 - 2)2
+ (2.5 - 0)2 + (2.5 - 8)2
+ (2.5 - 0)2] /
4 = 10.75
after
5 generations, when the number of Tonr cells has doubled in each culture:
variance = [(5.0 - 4)2
+ (5.0 - 0)2 + (5.0 - 16)2
+ (5.0 - 0)2] /
4 = 48.00
Mutation Hypothesis predicts variance
>> mean, as g increases
Experimental
procedure:
"The first experiment was done on
the
following Sunday morning.
(In a letter date January 21 [1943],
Delbruck exhorted me to go to church"
Twenty x
200 ul "individual cultures"
One x 10 ml
"bulk culture"
Inoculate with ~
103 bacteria @
Grow for g =
17 generations
=> ~108 bacteria / ml
Plate entire "individual cultures"
& 200 ul aliquots of "bulk culture" on dish w/ T1
Results
|
Individual
Cultures
|
Bulk
Cultures
|
|
Mean
|
11.3
|
16.7 |
|
Variance
|
694
|
15 |
In bulk culture,
a
= n / N = (16.7 / (0.2 ml x 108 bacteria / ml) = 8
x 10-7
variants / cell
variance ~
mean => random distribution
Expected result if
changes are induced (also compatible with mutation)
In individual culture
mean ~
mean in bulk
variance
>> variance in bulk:
Prediction of Mutation Hypothesis is supported
Mutation
rate (a) can be
calculated
mean # mutations / culture = aN
Poisson distribution predicts p0 = exp (- a /
N)
where p0 = fraction of cultures with no Tonr
mutants
Rewrite as a = (- ln p0) / N
and p0 = 11 / 20 = 0.55 from data
Then a = -ln 0.55 / (0.2 x 108)
= 3 x 10-8
mutations / cell / generation
Conclusions
"On a
postcard dated January
24, Delbruck replied:
|
January
24, 1943
From: Max
To: Salvador
"You are right about the difference
in fluctuations of resistants, when plating samples from one or from
several
cultures. In the latter case, the number of clones has a Poisson
distribution. I think what this problem needs is a worked out and written down
theory,
and I have begun doing so."
|
The MS of
the theory arrived on February
3...."
Luria on the significance of
these
experiments:
(1) "Adequate"
evidence of spontaneous mutation
rather
than induction
(2) Provided
method
for measuring mutation rates, and therefore is
(3)"The
Birth of Bacterial Genetics"
by showing that bacteria can be used to measure extremely low mutation
rates
All text material ©2005 by Steven M. Carr