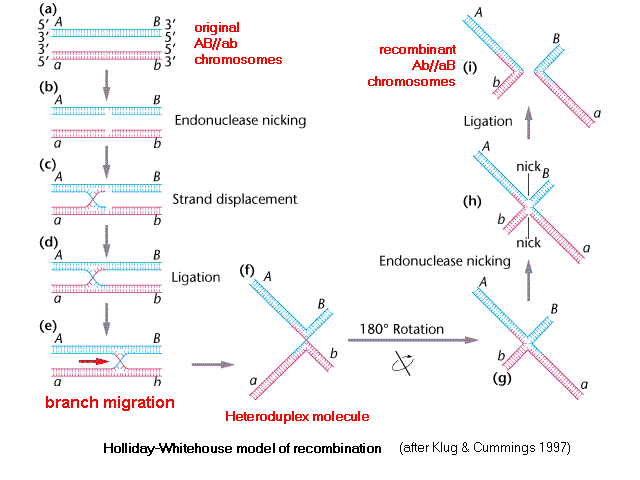
Holliday-Whitehouse Model of DNA Recombination
During Meiosis
I, (a) two homologous chromosomes line
up. Each shows a cis configuration of the A
& B loci, one is AB
and other ab , which may be
written AB
// ab.
(b) Single-stranded
nicking of one of the two DNA strands in each
chromosome provides an exposed 5' end on each
strand [trace this out] that
can (c) be displaced
or 'jump' to the other DNA molecule. (d)
Ligation of the
strands produces a covalently-joined heteroduplex branch
region, in which the bases are paired between opposite
chromosomes. (e) As the chromosomes are pulled
apart during Meiosis, the heteroduplex branch region
moves towards the opposite end of the pairing. [Think
of the centromeres as being out of sight, to
the left, such that the blue molecule is being pulled up
and the red molecule is being pulled down].
(f) When this is shown with the heteroduplex drawn
as an "X", the
heteroduplex region is clear, and when the "X" is rotated, (g)
the pattern of base-pairing in the heteroduplex is still
clearer. (h) Single-stranded
nicks in the non-recombinant chromatids from step (b)
separate the heteroduplex into two (i) recombinant molecules,
which are then ligated.
The genetic effect of the molecular mechanism is to reverse the linkage relationships (phases) of the alleles at loci on either side of the recombination event, so that the original cis configuration AB // ab has been changed to a trans configuration Ab // aB. If the markers were originally in trans, they would be reversed to cis.
The genetic effect of the molecular mechanism is to reverse the linkage relationships (phases) of the alleles at loci on either side of the recombination event, so that the original cis configuration AB // ab has been changed to a trans configuration Ab // aB. If the markers were originally in trans, they would be reversed to cis.
All text material ©2024 by Steven M. Carr