Bootstrap confidence with larger
vs smaller data sets


Bootstrap confidence with larger
vs smaller data sets
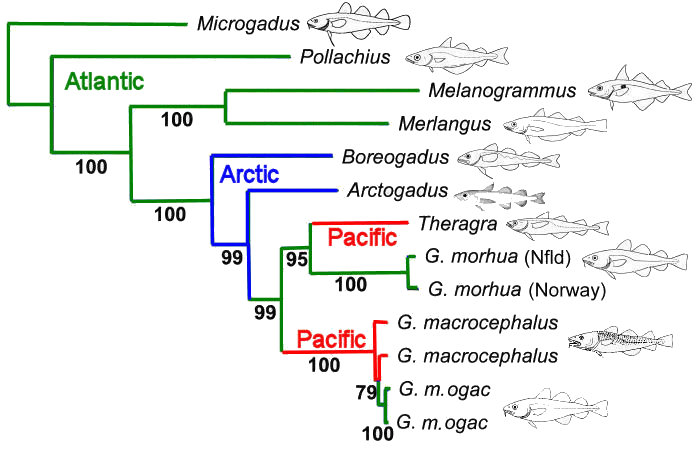
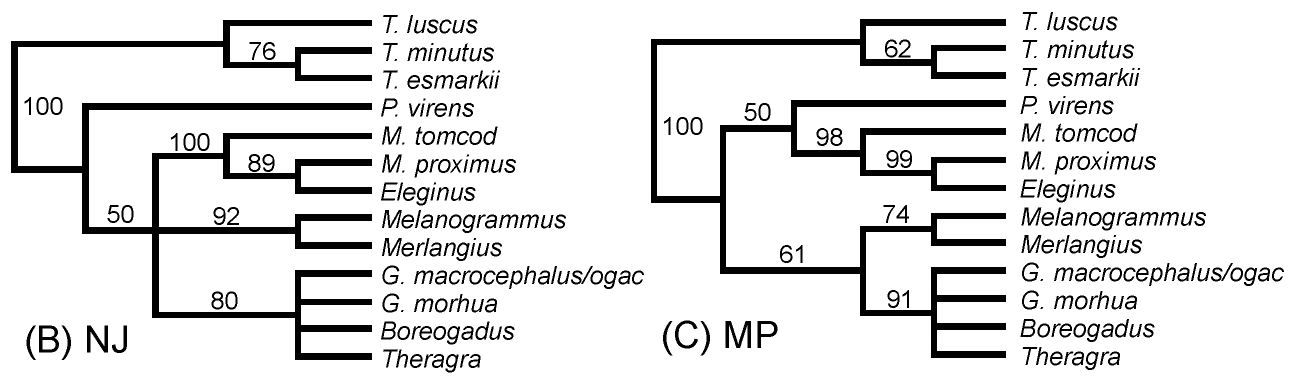
Evolutionary relationships among Gadid fishes (cods &
pollocks) were analyzed first with partial (0.8 Kbp, Diagrams B
& C, below) and later with complete mtDNA genomes (16.0 Kbp, above).
The 20-fold increase in data allows resolution of
phylogenetic relationships by the bootstrap with greater
confidence. Note that all but one of the within- &
among-species relationships in the upper tree are resolved at >95%
confidence, whereas a minority of branches are so resolved in
the smaller data set.
Resolution of the
branching order shows that (1) the two Pacific Gadid
species, Pacific Cod (Gadus macrocephalus) and Alaska
Pollock (Theragra chalcogramma) are separate
invasions of the Pacific basin, (2) the latter should be
included in Gadus as Gadus chalcogrammus,
rather than placed in a separate genus, and (3) Greenland
cod (G. ogac) are genetically almost identical to Pacific
Cod (G. macrocephalus) despite their geographic
separation, which suggests a relatively recent re-invasion
of the Arctic / Atlantic basin by the former.
B & C: 0.8
Kbp analysis from Carr et
al. 1999, by Neighbor-Joining and Maximum
Parsimony methods;
(A) 16.0 Kbp Whole-genome analysis from Coulson et al. 2006