Exons, Introns, Codons, & their equivalents
Three common technical terms in molecular genetics, exon, intron, and codon, have specific technical definitions, but are often miss-used in hurried or short-hand presentations. Exons and introns are features of DNA; codons are features of mRNA. Homologous sequences in the other type of nucleic need to be called something else, otherwise there is a danger the roles of DNA and RNA in the Central Dogma ("DNA makes RNA makes Protein") will be confused.
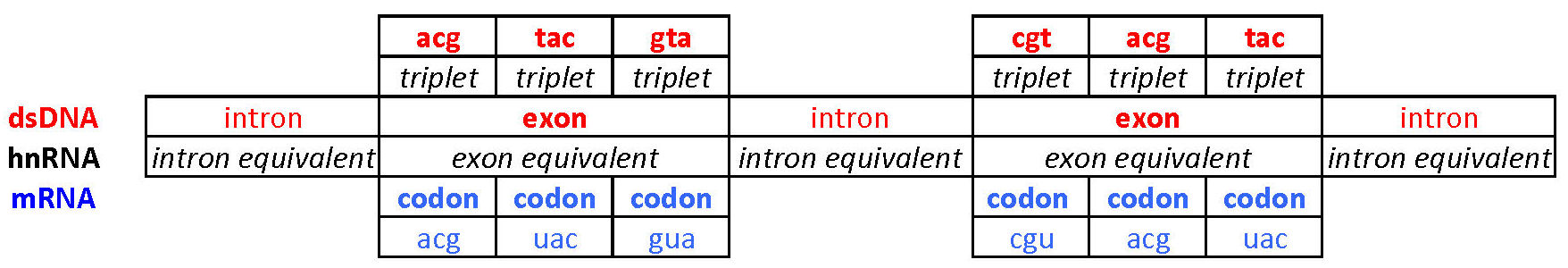
By definition, exons and introns are sequences in a protein-coding gene region of a double-stranded DNA molecule (dsDNA) that are expressed as proteins, or intervening sequences not so expressed. The exons and introns are typically shown as the single-stranded sequences of the Sense Strand of the dsDNA, written 5'-3', left to right.
Transcription of the complementary Template Strand produces a heterogeneous nuclear RNA (hnRNA) that is identical (co-linear) in 5'-3' orientation and base sequences to the DNA Sense Strand, with the substitution of U for T. The RNA sequences equivalent to the DNA exons and introns are sometimes themselves referred to as "exons" and "introns." However, this is technically incorrect, and also confuses their functional role in transcription and translation with their informational role as structures in the gene sequences in DNA. The RNA sequences equivalent to to DNA exons and introns should be referred to as "exon transcripts" and "intron transcripts," or "equivalents," respectively.
Processing of the hnRNA to mRNA involves excision ('splicing out') of the intron transcripts and ligation of the remaining exons. Once the final mRNA is formed, translation is the process of reading (as amino acids) a series of three-base sequences called codons. Codons are read according to the Genetic Code, which is an RNA code. Because the mRNA region is equivalent to a DNA exon, the same three-base series can be identified in the Sense Strand (substituting T for U). The three-base DNA motifs are some called "codons", however this is again technically incorrect and confuses the information content of Genes with the function of RNA in the Genetic Code. The DNA equivalents to codons can be referred to as 'triplets.'
In bioinformatics, the 64 triplets are sometimes presented as a "translation table" that can be used directly with the DNA Sense Strand sequence to infer the protein sequence. This is practical, except that "translation" here means 'deciphering coded information', which is not the same as the molecular process of mRNA translation.
There's an app for that: see SM Carr, HT Wareham & Craig D. 2014. A web application for generation of DNA sequence exemplars with open and closed reading frames in genetics and bioinformatics education. CBE – Life Sciences Education 13, 373-374, which reviews this and includes an app that renders dsDNA as protein sequences.
Figure & Text © 2024 by Steven M. Carr