RNA sequence evolution in the
COVID-19 SARS rRNA virus
The RNA Corona virus responsible for COVID-19
SARS (Severe Acute Respiratory Syndrome) first
emerged in the vicinity of Wuhan, China, in late 2019 (hence the '-19').
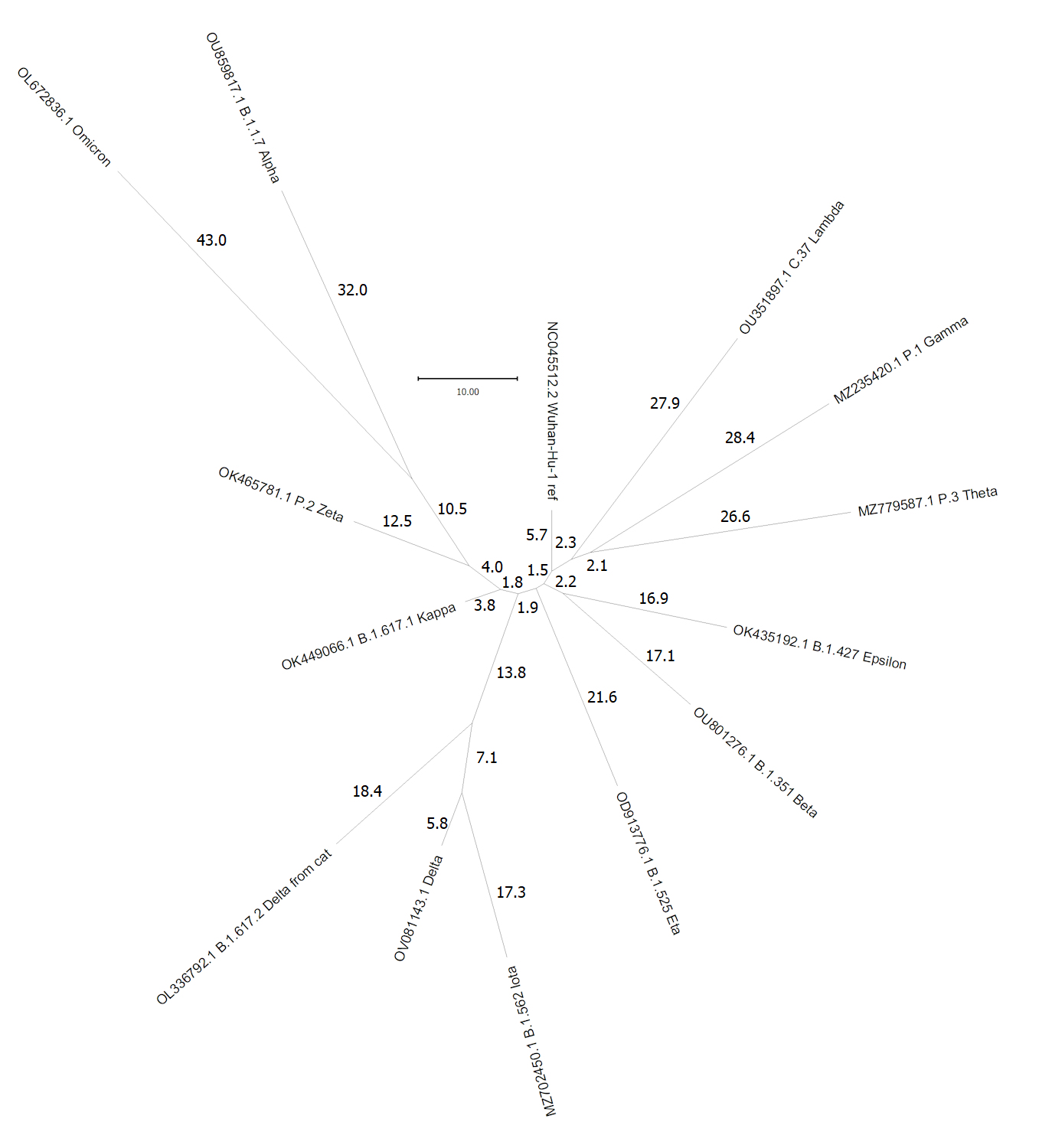
Its RNA sequence comprises about 30,000 A C G & U
s, and Wuhan-Hu-1 [top, at 12 o'clock]
is used as the reference for comparison with new strains. The
sequence is catalogued in NIH's NCBI (National
Center for Biotechnology Information) on-line
GenBank library
as accession NC045512.2.
NCBI maintains the Virus
Data Hub, which collates new sequence data as they
emerge.
The original Wuhan-Hu-1 virus
sequence has mutated (A C G U bases in the
sequence change to different letters) as it has spread through the
human population. Virus mutation is extremely rapid: related lineages
of the virus can be identified by shared mutations
between isolates collected at different times and places, in the
same way that DNA sequences are used to track family
trees in humans. The diagram reconstructs the evolutionary
relationships among some of these lineages through 2021. Numbers
on each of the branches indicate the number of mutations that have
occurred. For example, the "Iota" variant of the "Delta"
strain [bottom, 7 o'clock] differs from it by (17.3 + 5.8) = 23.1
mutations. Greek letter names are assigned to major new strains,
especially when its mutations affect its biology. For example,
gene mutations in the Delta strain lead to changes in the
critical 'Spike Protein' that affect how the virus
attaches to human cells, and therefore affect its infectivity and
transmission. Another Delta variant shown was isolated
from a house cat in France: animal populations can act as
reservoirs for viruses that produce symptoms only in humans. The
original Wuhan virus appears to have arisen as a hybrid between
bat and humans viruses.
Lineages of present concern are the Delta variant
[bottom, 7 o'clock], which has a higher infection rate than
previous forms, and the Omicron variant [10 o'clock],
which emerged in November 2021 in southern Africa and has rapidly
spread to other populations. Omicron is notable for
having an extremely large number of mutational differences from
the Wuhan reference (~ 70), many of which occur in the 'Spike
Protein' gene, and have the potential to change the
way in the which the virus attacks human cells, and (or) the
ability of existing vaccines to guard against it.
Figure & Text material ©2021 by Steven M. Carr; not to be
reproduced without permission

