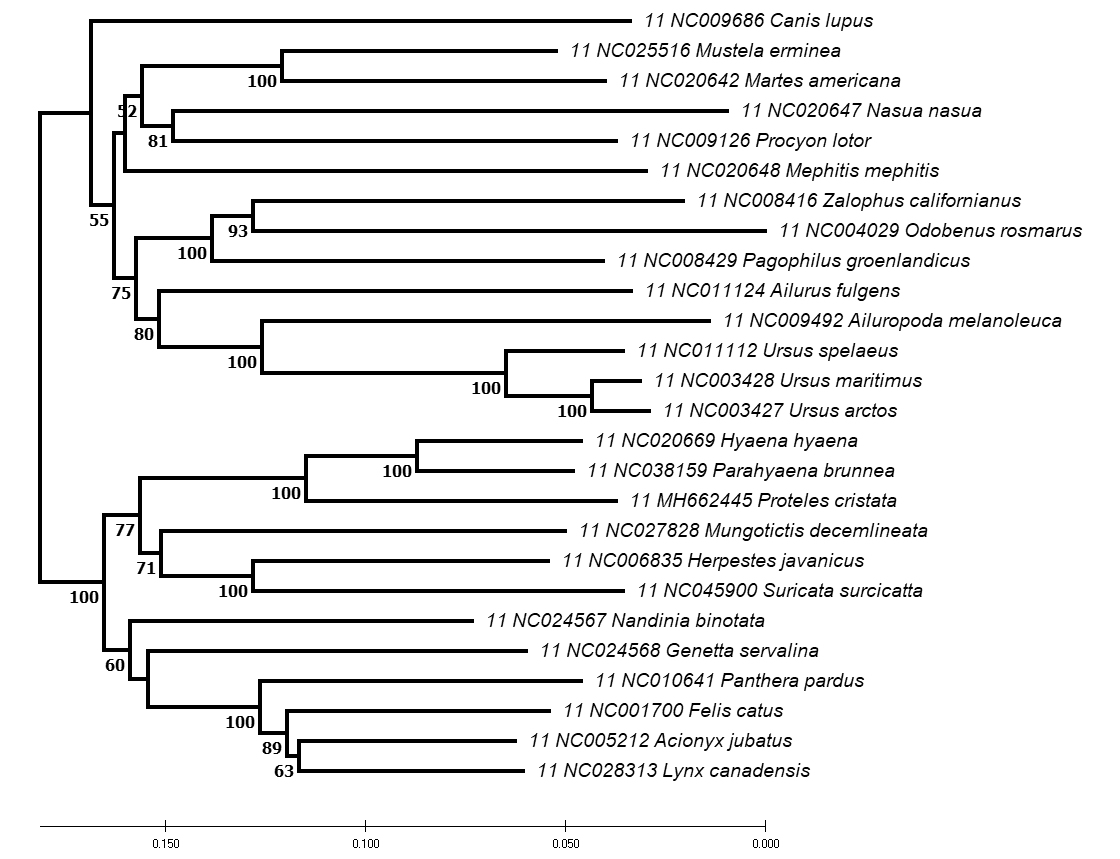
Evolutionary
relationships among 26 species of Carnivora
based on 2,973 bp of mtDNA ND5
& Cytb genes

Evolutionary
relationships among 26 species of Carnivora
based on 2,973 bp of mtDNA ND5
& Cytb genes
The phylogenetic
tree above is based on a Neighbor-Joining Analysis in
MEGA of 2,973 bp of the mtDNA ND5 (1,833 bp)
and Cytb (1,140bp) genes. Numbers in BOLD below
each node indicate the percent bootstrap support from
3,000 bootstrap replications. Bootstrap values < 50% are not
shown.
The network has
been rooted to separate the dog-like Caniformia species
(above) from the cat-like Feliformia species (below).
Species arrangements within each suborder are unmodified from
the MEGA output.
QUESTIONS: The tree allows molecular inferences about relationships among Carnivore families.
1. According to
the tree, are the following families as recognized by
conventional morphology-based taxonomy Monophyletic OR
Paraphyletic (= Polyphyletic)? State your
reasoning as supported by the tree.
a. Felidae
b. Mustelidae
c. Hyaenidae
d. Viverridae
2. Is the Giant
Panda more closely related to bears or to the Lesser
Panda? State your reasoning as supported by
the tree.
3. Are the marine mammals (Pinnipedia)
monophyletic or Polyphyletic? State your
reasoning as supported by the tree.