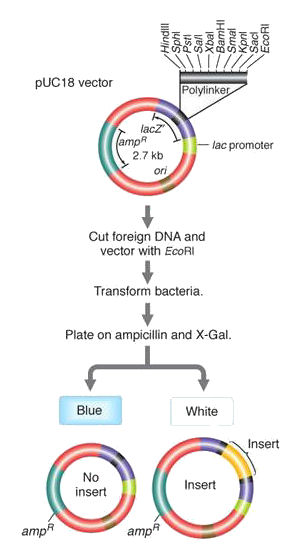

Selection scheme for recombinant plasmids
The plasmid vector
includes (1) an ampR gene for
resistance to the antibiotic ampicillin,
(2) a polylinker region
that contains a number of unique restriction endonuclease
recognition sites, located inside a (3) LacZ gene
that allows the plasmid to metabolize the sugar X-galactose (X-Gal) and produce a blue
by-product.
A foreign DNA is restricted with one of the endonucleases in the polylinker, and recombined with a linearized plasmid cut with the same enzyme. The plasmids are allowed to transform a population of bacteria. Of the very large number of bacteria in the experiment (106~9s), only a small fraction take up a plasmid, and of these only a few of those plasmid contain recombinant DNA. Rather than screen millions of bacteria individually, a two-stage selection scheme is employed to screen out, first untransformed bacteria, and then those without recombinant plasmids.
The bacteria are grown on a petri dish with ampicillin and X-galactose. (1) Only those cells that have an ampR gene from the plasmid can grow at all. (2) Of those that took up a plasmid, those that do not contain recombinant DNA have an intact polylinker in the lacZ gene and are thus able to metabolize X-Gal, Alternatively, successful insertion of recombinant DNA into the polylinker disrupts the lacZ gene, such that it is non-functional. Blue and White colonies thus signal unsuccessful and successful creation of recombinant DNA, respectively.
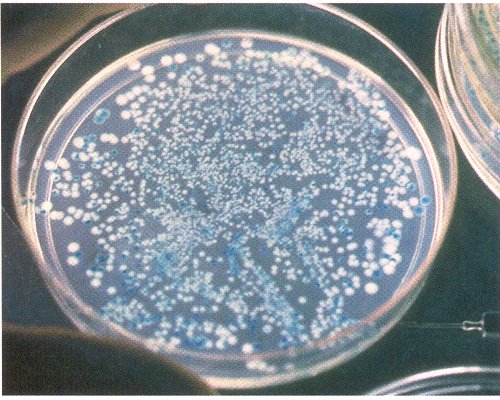
On the petri plate at right, multiple millions of bacteria do not grow at all, a few hundred blue colonies grow without recombinant inserts, and only a few dozen white colonies indicate successful uptake of foreign DNA. Subsequent analysis will focus on these.
A foreign DNA is restricted with one of the endonucleases in the polylinker, and recombined with a linearized plasmid cut with the same enzyme. The plasmids are allowed to transform a population of bacteria. Of the very large number of bacteria in the experiment (106~9s), only a small fraction take up a plasmid, and of these only a few of those plasmid contain recombinant DNA. Rather than screen millions of bacteria individually, a two-stage selection scheme is employed to screen out, first untransformed bacteria, and then those without recombinant plasmids.
The bacteria are grown on a petri dish with ampicillin and X-galactose. (1) Only those cells that have an ampR gene from the plasmid can grow at all. (2) Of those that took up a plasmid, those that do not contain recombinant DNA have an intact polylinker in the lacZ gene and are thus able to metabolize X-Gal, Alternatively, successful insertion of recombinant DNA into the polylinker disrupts the lacZ gene, such that it is non-functional. Blue and White colonies thus signal unsuccessful and successful creation of recombinant DNA, respectively.
On the petri plate at right, multiple millions of bacteria do not grow at all, a few hundred blue colonies grow without recombinant inserts, and only a few dozen white colonies indicate successful uptake of foreign DNA. Subsequent analysis will focus on these.