Assessment of Phylogenetically Informative nucleotide sites

Assessment of Phylogenetically Informative nucleotide sites
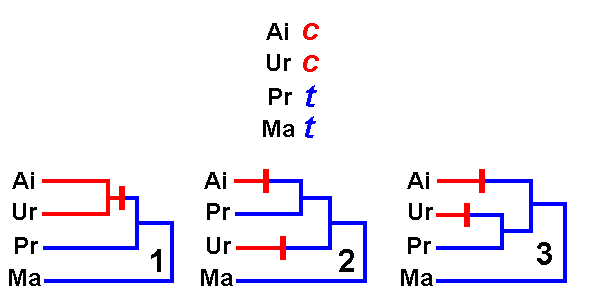
We are interested in relationships among the Giant Panda (Ailuropoda), a bear (Ursus: Ursidae), and a raccoon (Procyon: Procyonidae). We include a fourth species, the pine marten (Martes: Mustelidae), as an outgroup known to be a distant relative of the first three. There are three possible hypotheses of evolutionary relationships among these species: either (1) Ailuropoda is most closely related to Ursus, or (2) Ailuropoda is most closely related to Procyon, or (3) Ursus & Procyon are each other's closest relatives.
Given a set of homologous DNA sequences
from these four species, we are looking for positions at which two species
share a particular nucleotide, and the other two species share a
different nucleotide. Such sites are said to be phylogenetically informative,
because they favor one hypothesis of relationships over the others.
In the example, Ailuropoda &
Ursus are both c
at a particular position, and Procyon and Martes are both
t
at the same position. Positions of the type shown can be explained in the
first hypothesis by a single mutation from t![]() c
that occurred in the common ancestor of Ai & Ur. Either
of the two competing hypotheses require two mutations to explain
this position. Therefore, the first hypothesis is a more parsimonious
(simpler)
explanation of the data than is either of the others.
c
that occurred in the common ancestor of Ai & Ur. Either
of the two competing hypotheses require two mutations to explain
this position. Therefore, the first hypothesis is a more parsimonious
(simpler)
explanation of the data than is either of the others.
The cladistic DNA analysis counts the number of informative sites that favor each of the three hypotheses. The hypothesis favored by the greatest number of such sites will require the fewest mutations [be sure you understand this!] and is therefore the most parsimonous solution overall.