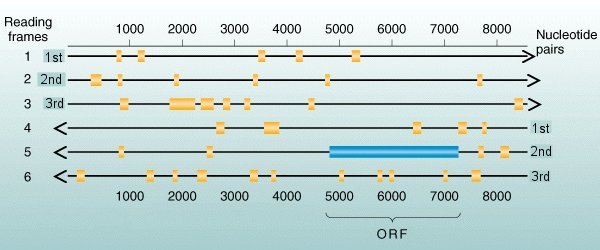
Any piece
of dsDNA can be read in six possible ways. The
diagram shows a single dsDNA molecule: either strand
could be the sense strand read in the 5' ![]() 3'
direction, and on either possible sense strand there are three
possible start points, beginning at the 1st, 2nd,
or 3rd nucleotides. Each of these 2 x 3 = 6
possibilities is called a reading frame.
3'
direction, and on either possible sense strand there are three
possible start points, beginning at the 1st, 2nd,
or 3rd nucleotides. Each of these 2 x 3 = 6
possibilities is called a reading frame.
Because
three of the 64 possible DNA
triplets correspond to
mRNA stop codons, a DNA sequence read at random will have stop triplets approximately once in
every 20 triplets. The occurrence of multiple stops in a
particular reading frame indicates that it does not code for a
polypeptide: this is a "closed"
reading frame. In contrast, an Open Reading Frame (ORF) can be read through
several hundred triplets without encountering a stop sequence.
ORFs are therefore candidates for protein-coding exon regions: the inferred amino-acid sequence
can be compared with GenBank
to identify possible analogous proteins.
In the
example above, the longest open runs in reading frames ##1, 2, 3, 4, & 6 [gold boxes] are
very short runs, typically < 100 bases of amino acids
without stops. Reading
frame #5 , read 5'![]() 3' right to left, includes
an ORF [blue box] of
more than 2,000 nucleotides, corresponding to a protein of
more than 600 amino acids.
3' right to left, includes
an ORF [blue box] of
more than 2,000 nucleotides, corresponding to a protein of
more than 600 amino acids.
Figure ©2000 by Griffiths et al. ; text ©2014 by Steven M. Carr