Any piece
of dsDNA
can be
“read”
in six possible ways. There are two strands,
either of
which could be the sense strand read in the 5' ![]() 3'
direction, and on
either possible sense strand there are three possible
start points, beginning
at the 1st,
2nd, or 3rd nucleotides. Each of
these
six possibilities is called a reading
frame.
3'
direction, and on
either possible sense strand there are three possible
start points, beginning
at the 1st,
2nd, or 3rd nucleotides. Each of
these
six possibilities is called a reading
frame.
Because 3
of
the 64 possible
DNA triplets
correspond
to mRNA stop codons, a DNA
sequence read at random will
have 3-letter stop signals
approximately
once in every 20 triplets. The occurrence of
multiple stops in a particular reading frame indicates that it
does not
code for a
polypeptide: this is a "closed"
reading frame. In contrast, an Open
Reading
Frame
(ORF) can be read
through several
hundred triplets without encountering a
stop
sequence. ORFs are therefore candidates for
protein-coding exon regions: the
inferred amino-acid sequence can be compared with GenBank
to identify possible analogous proteins.
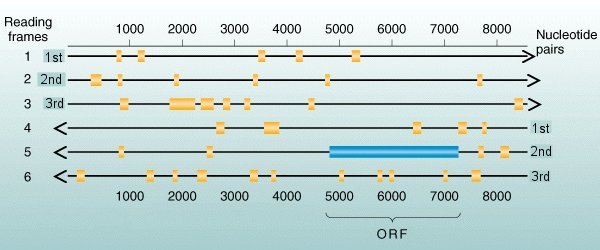
In the
example
above, the longest open runs in reading
frames ##1, 2, 3, 4,
& 6 [gold boxes] are very short
runs, typically < 100
bases of amino acids without stops. Reading
frame #5 , read 5'![]() 3' right to
left, includes an ORF
[blue box]
of more than 2,000 nucleotides, corresponding to a protein of
more than
600
amino acids.
3' right to
left, includes an ORF
[blue box]
of more than 2,000 nucleotides, corresponding to a protein of
more than
600
amino acids.
Figure © 2000 by Griffiths et al. ; text © 2012 by Steven M. Carr