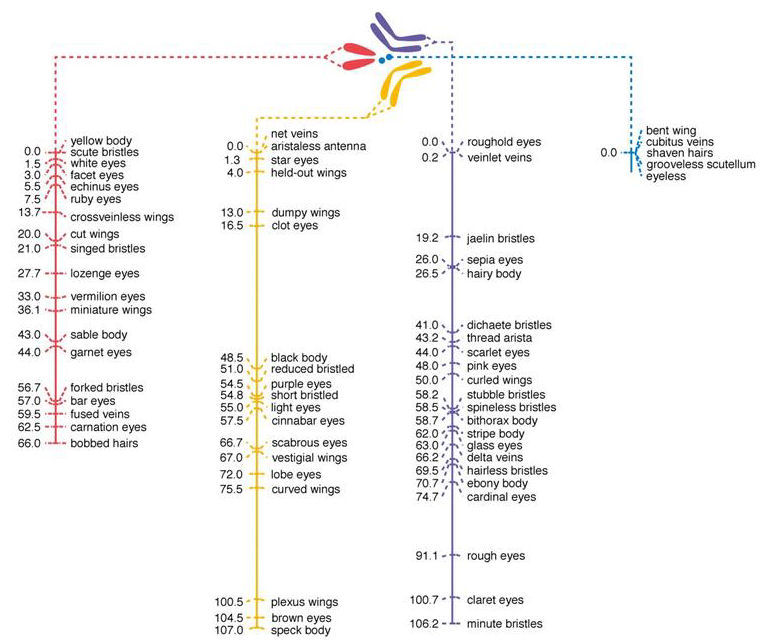
Genetic linkage map of the Fruit Fly, Drosophila melanogaster (2n = 4)
The first gene maps were drawn in 1916 from
recombination frequency data collected by Calvin
Bridges (1889-1938), a student of Thomas Hunt
Morgan. Dihybrid crosses between flies
typically followed the Mendelian expectation of independent
assortment of genes. However, for some pairs,
fewer recombinant types than expected were obtained.
Bridges reasoned that the recombination frequency
might provide an estimate of the physical distance
between the genes on a chromosome. Bridges called a 1%
recombination frequency 1 centimorgan (cM)
in honor of his boss. By constructing crosses between
flies that differed at three markers, the relatives
order of genes could also be determined. Bridges also
realized that the genes fell into four linkage
groups, corresponding to the four chromosomes
known from cytology, thus proving the chromosome
theory of heredity. Genes in different linkage groups
always segregated independently, whereas genes
in any one linkage group segregate together
depending on the distance between them.
Extensive physical mapping of Drosophila for dozens of classical gene variants provided a highly resolved map for early localization of these genes on DNA molecules, and eventually a scaffold on which to assemble multiple overlapping DNA sequences from many thousands of DNA fragments. The first genome DNA sequence of a complex animal was obtained for Drosophila in March 2000.
Extensive physical mapping of Drosophila for dozens of classical gene variants provided a highly resolved map for early localization of these genes on DNA molecules, and eventually a scaffold on which to assemble multiple overlapping DNA sequences from many thousands of DNA fragments. The first genome DNA sequence of a complex animal was obtained for Drosophila in March 2000.