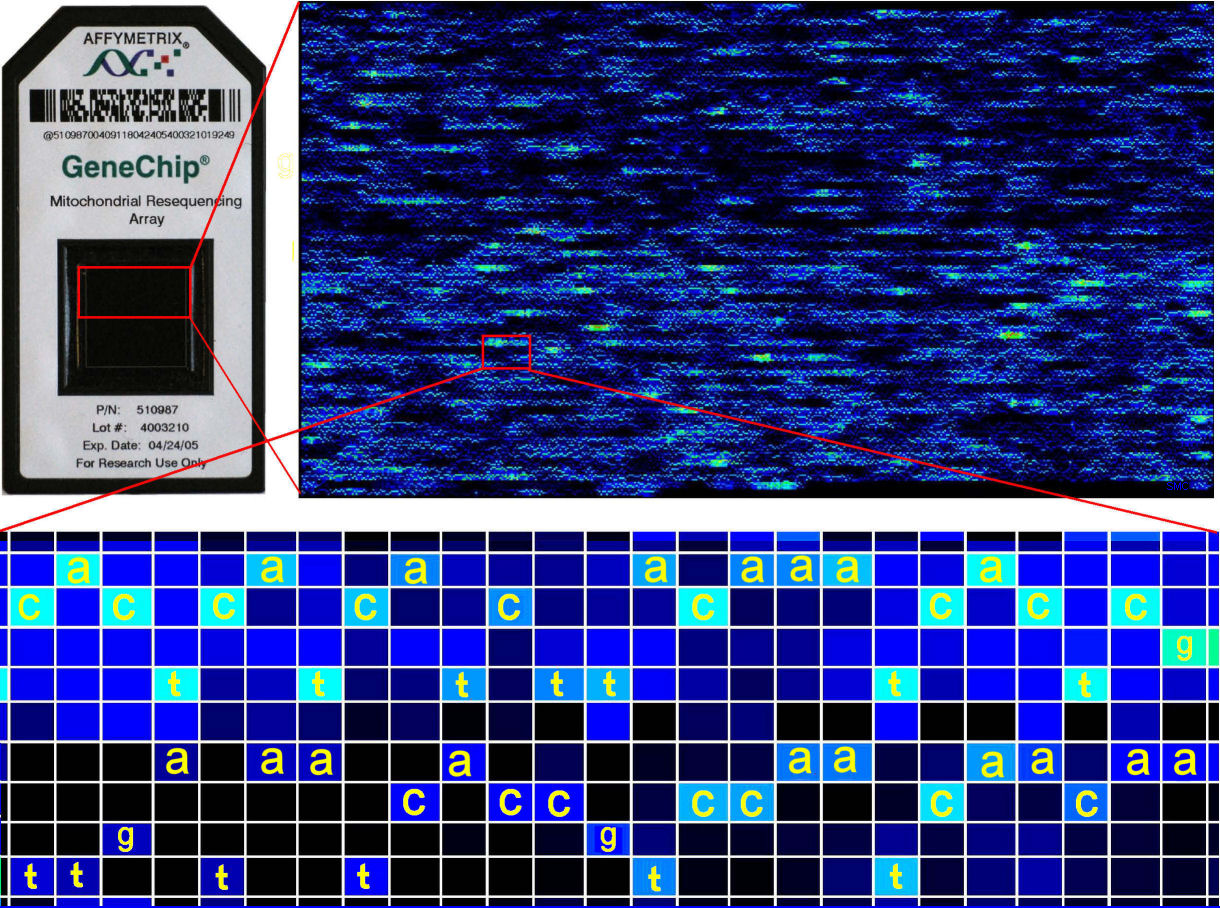
Alternative approaches
to SNP detection:
Iterative DNA "Re-Sequencing" on Microarrays
(after SM Carr et al. 2008 Comp Biochem Physiol D, Genomics &
Proteomics 3,1-11)
A DNA
Re-sequencing microarray is based on a reference sequence of
length n bases, represented in a
series of n overlapping ("tiled")
oligonucleotide probes. For each probe, three variant probes are
included that vary the middle base, one for each of the three
alternative DNA code letters.
Mis-match at this position most strongly influences binding, so that a
genomic DNA fragment
with a SNP
sequence
will stick
to only one of the four oligos at any tiled position.
Re-sequencing allows a lengthy contiguous sequence to be determined in
one
experiment, without multiple PCR and sequencing reactions. Re-sequencing
of both strand of a
continuous 17Kb sequence like mtDNA requires 2 x 4 x 17,000 = 136,000 oligos [above]. Re-sequencing is useful where analogous
sequences
are to be read
repeatedly ("re-sequenced")
from multiple individuals, and is thus particularly well-suited to population genomics.
In the enlargement below, the re-sequencing chip
tiles the reference sequence in
the first row, and the variants
below it. (A vs CGT,
C vs AGT
, G vs AGT, and T vs ACG). The
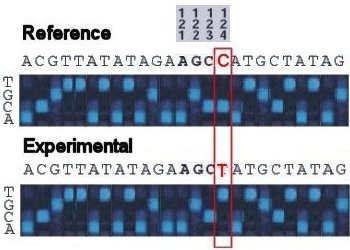
occassional variant stands out clearly: the reference DNA sequence is AGCC at positions 121-124, and the experimental
sequence is AGCT.