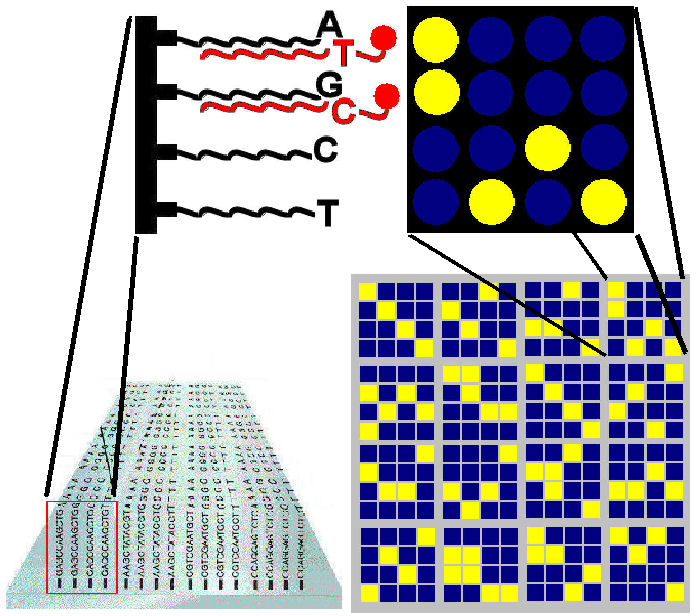
Principle of a DNA microarray chip: use as Variant Detector Arrays (VDAs)
(after SM Carr et al. 2008. Comp Biochem Physiol D, 3:11)

A DNA chip is a small piece of
silicon glass (~1 cm2) to which a large number of
synthetic, single-stranded DNA oligonucleotides ("oligos") have
been chemically bonded [left]. Oligos function as DNA probes: they "stick" (anneal) selectively
only to those DNA molecules
whose nucleotide sequences are exactly complementary: T pairs with A, and G with C. They can therefore be
used to identify the presence
of specific DNA sequences in a heterogeneous mixture of
genes, for example the presence of a particular allele against
the background of a complete genome. In effect, oligos act like
molecular "velcro." A
computer "reads" the
pattern of annealing and "reports"
which alleles are present.
DNA chips can be used s Variant Detector Arrays (VDAs)
to look for DNA sequences
that differ by single
nucleotide polymorphisms ("SNPs").
In this example, the DNA sequences
of the four oligos
highlighted in the first bloc differ only at the last position.
To determine which alleles are present, genomic DNA from an
individual is isolated, fragmented, tagged with a fluorescent
dye, and applied to the chip. The genomic DNA fragments
anneal only to those oligos to which they are perfectly
complementary: in this case, the allele with the ~T~ SNP allele binds to the ~~A
oligo, and the allele
with the ~C~ SNP
allele binds to the ~~G
oligo. A computer reads the position of the two
fluorescent tags and identifies the individual as a C / T heterozygote.
[The single spots in
the other three columns indicate that the individual is homozygous at the three
corresponding SNP positions].
The 4 x 4 arrays fits in one corner of a 256-oligo chip (lower right). The current generation of microarrays can accommodate hundreds of thousands of oligos.