Reference: Griffiths et al. Modern Genetic Analysis, 2nd Edition 2003
Chapter 18
Quantitative Genetics - The study of genetics of continuosly varying characteristics- Variations in most populations consist of a continuous phenotypic range instead of discrete phenotypic classes (ie: complete blue or green eyes and partial amounts of blue and green)
Quantitave versus Qualitative
Mendelian Genetics
- Mendelian Genetics applys qualitative measurements and is not useful when dealing with continuous phenotypic range
THEREFORE
Statistical techniques and analysis are
applied to measure these variations
Examples of Statistical Measures Used in Quantitative Genetics
Multiple factor hypothesis - describes that a large number of genes, each with a small effect, are segregating to produce quantitative variation.
Example: Wilhelm Johannsen corn experiment
Johannsen used pure line ( family lines that are true breeding from generation to generation) in his quantitative genetics analysis experiment.
He produced these true lines of corn by inbreeding and selfing.
He hypothesized that each subsequent generation would exhibit the same phenotypic expression. He calculated the number of expression by the formula :
Heritability of a Trait
- Question of the day about a quantitative trait!!!
- Every character has a gene influenced developmental process within it, but variation is not neccesarially the results of genetic varaition.
- Most traits are polygenic (influenced by more than one gene)
- Traits that are familial are when members of the same family share them
- Traits are heritable when the similarity arises from shared genotypes
Two general methods of finding the heritability of a trait
- Depends on phenotypic similarity between relatives
- Uses marker-gene segregation
Ideal subjects are identical twins
Quantifying Heritability
Phenotypic variance (Sp^2) can be broken into two parts- Genetic variance (Sg^2) - variance between genotypic
means
- Enviromental variance (Se^2)
Problem: Excludes co-variance between genotype and enviroment
Example: Musical parents rearing musical children - Genetics versus Enviroment ???
Therefore, essential to include co-variance: Sp^2 = Sg^2 + Se^2 + 2 cov ge
Broad heritability (H^2): the degree of heritability of a character
Method of estimating H^2
- Makes use of genetic similarity between siblings
- Genetic correlation for siblings = 1/2
- For half siblings = 1/4
- Therfore, the difference in genetic correlation between full and half sibs is 1/4
- Have to include enviromental variance, so use phenotypic correlation
Can also use parent or twin inforamtion
Meaning H^2
Two conclusions from heritability studies
- If H^2 is non-zero, genetic differences have influenced variation between individuals, if 0, does not neccesarily mean genes are relevant
- H^2 is limited in prediction of effects of enviromental modification under particular circumstances
Locating the Genes
Not possible with genetic techniques to identify all genes that influence the development of a given trait.Genetic Analysis - detects only when there is some allelic variation
Molecular Analysis - can identify genes even when they do not vary (provided the gene products can be identified
A trait may show continuous phenotypic variation, but the genetic basis for the differences must be an allelic variation at a single locus.
It is possible to use prior knowledge of biochemistry and development to guess that variation at a known locus is responsible for at least some of the variation in phenotype.
This locus is then a candidate gene for investigation of continuous phenotypic variation
Marker Gene Segregation
- Quantitative Trait Loci (QLTs) = genes segregating for a quantitative trait
- Cannot be individually identified in most cases
- Possible to localize those regions of the genome in which the relevant loci lie
- Analysis is done in experimental organisms by crossing two lines that differ markedly in the quantitative trait and differ in alleles at known loci, Marker Genes
- Has to be where different genotypes can be distinguished by criteria such as some visible phenotypic effect (ie. Drosophilia eye color)
- Localization of QLTs to small regions within a chromosome requires that there be closely spaced markers along the chromosome
- An unlinked marker locus will have the same average value of the quantitative trait for all its genotypes
- For humans:
- Possible to use the differences among sibs carrying different marker alleles from heterozygous parents. This method has much less power to find QLTs.
- Consequence - attempts to map QLTs for human traits have not been succesful
- Although, segregating marker techniques has been a success in finding loci whose mutations are responsible for 1 gene disorder
More analysis of variance
Two Types
- Additive Variance (Sa^2) - The genetic variance associated with the average effects of substituting one allele for another
- Dominance Variance (Sd^2) – The genetic variance at a single locus that is attributable to dominance of one allele over another
h^2 – proportion of phenotypic variance accounted for by additive genetic variance only (heritability in a narrow sense). This value is useful in determining whether a program of selective breeding will succeed in changing a population.
h^2 = Sa^2 / Sp^2
The effect of selection depends on the amount of additive genetic variance in a population and not on the genetic variance in general. Therefore, the narrow heritability, h2, not the broad heritability H2, is relevant for a prediction of response to selection.
Estimating the Components of Genetic Variance
• All non additive variance is attributed to dominance variance
• The slope of a regression line in comparing offspring values to midparent values for a character with heritability can be used to estimate h2. This can then be used to predict the effects of artificial selection (figure 18-14)
Midparent value – average phenotype of two parents
Selection Response – difference between the offspring of the selected parents and mean of the parental generation
Selection differential – the difference between the mean of a population and the mean of the individuals selected to be parents of the next generation.
Selection Response = h2 * Selection Differential
• h2 estimate depends on the assumption that there is no correlation between similarity of individuals’ environments and the similarity of their phenotypes
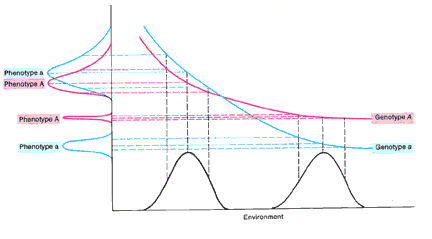
• h2 in one population in one set of environments will not be the same as h2 in a different population in a different set of environments (figure 18-13)
Use of h2 in Breeding
• Higher value of h2 reflects a higher offspring-parent correlation in heritability
• If h2 is low, then only a small fraction of parental superiority will appear in the next generation
• If h2 and H2 are both low, then this signifies a large proportion of genetic variance
• If h2 is low, but H2 is high, then there is not much environmental variance
Hybrid – Inbred Method – Large number of inbred lines from selfing. Lines are then crossed in many different combinations, and the cross that gives the best hybrid is chosen
The subdivision of genetic variation and environmental variation provides important information about gene action that can be used in plant and animal breeding.