Midterm Examination questions, Winter
2016 [Final Set, posted 22 Feb 2016]
Instructions: Prepare answers for all questions. FOUR of these
questions will be chosen at random for the midterm. Your may
answer ANY TWO of these. You MAY NOT answer the question based
on your group’s presentation. Answers should be no longer than
one side of an 8.5x11 sheet; you may sketch an outline on the
reverse. Answers will be graded on the expectation that you
have written out answers to all questions ahead of time, and
that the examination paper is only a check that this has been
done.
1.
Radiation Genetics & Health
Physics: Consider a lab accident involving 10 uCi
of a gamma- versus an alpha-emitter.
Which would you rather be exposed to at 1
meter? Which would you rather swallow? Explain
your reasoning..
3.
The Hershey & Chase (1952) experiment
has been criticized as being "quick and
dirty". In Figure
1 of the experiment, what result would you
expect to obtain if all phage protein remained outside,
and all phage nucleic acid went inside the bacterial cells?
What are some explanations
4.
Watson and (or) Wilkins have been
criticized for miss-use of data from Franklin. Based on the evidence
presented in lecture, to what extent is the criticism
justified? Give the facts and explain your reasoning. Which is
more critical to understanding the genetic properties of DNA,
its double helical structure or specific
base-pairing? Explain your reasoning.
5.
Meselson & Stahl (1958) use analytical
centrifugation to demonstrate the semi-conservative mode of
DNA replication. Sketch the banding patterns that would be
expected if DNA replication were conservative,
or dispersive. At what stage of the original
experiment can the conservative and dispersive models
be ruled out? Explain.
6.
McClintock
(1953) cross analysis: Based on the genetics of
the Ac-Ds system, explain the phenotypes of
the three kernels in Table 1.
7.
King & Wilson (1975) calculated from an electrophoretic
similarity of S' = 0.52 that chimps and
humans were >99% genetically similar. (1) Suppose
instead the data set included 20 loci that share an identical
allele between species, and 20 loci at which each
species has a separate allele. Summarize the data
table. (2) Calculate S. (3) Calculate the genetic
similarity between species; show your calculations.
8.
Nirenberg et al.
(1965) deciphered
the first four codons of the genetic code by use of poly
U, C, A, and G,
and a further set by use of all possible dinucleotide
combinations (UA, UC, UG,
CA, CG, AG, etc).
(1) Show which additional codons can be deciphered by the
use of dinucleotide polymer messages. What does
this information indicate about degeneracy of the code?
(2) Consider a poly-U message ‘spiked’ with
small amounts of A, or C, or G:
what additional codons, and what information about
degeneracy would be shown?
9.
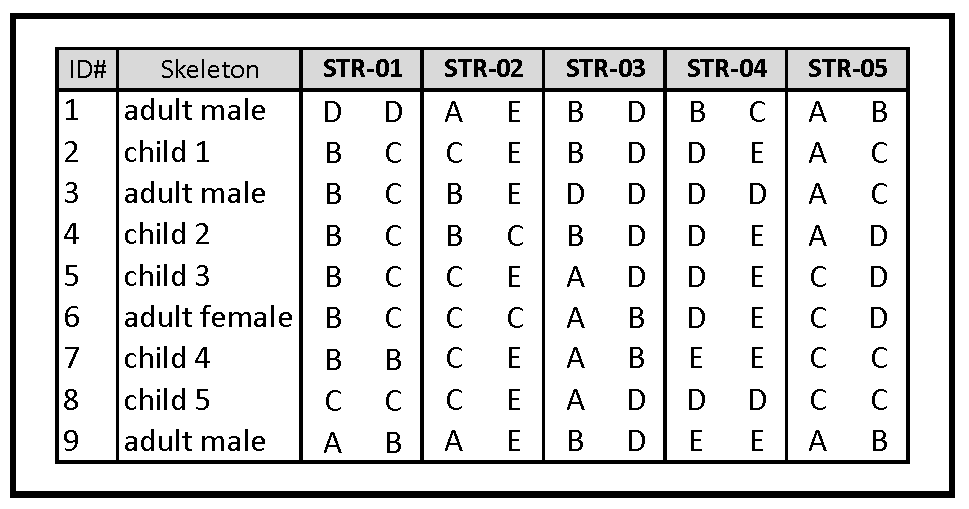
Gill et al. (1994): Consider an alternate
scenario for recovery of the remains of the Romanov family,
given below. The remains
include five children, a single adult female, and three adult
males. It is assumed that these include the Tsar, Tsarina, all
five of the children, and two males associated with the
family. Five STR loci were examined, each with four alleles
designated A-E as necessary.
1) Identify the Tsar. Show that the
STR genotypes of each child is consistent with parentage by
the Tsar an Tsarina.
2) Show that the other two males are not
the Tsar. Explain your reasoning.
3) Notice anything unusual? Explain
Hint:
For any two parents, there are a maximum of four (A,B,C,D) and
a minimum of one (A) alleles segregating at any locus. There
are also five intermediate configurations of two- (A,B,C) and
three- (A,B) allele parental genotypes. Writing out these
seven combinations, and the types of offspring each can
produce, may assist.

Text material ©2016 by Steven M Carr