Protein Structure & Function
In principle:
Proteins are polymers of amino
acids
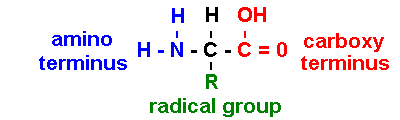
[sometimes NH3+ & COO- : depends on pH] [iG1 7.02]
R = radical group:
asymmetric (); levo
(L)-rotatory
[cf. dextro (D)-rotatory]
determines biological properties: 20 types (note 1- & 3-letter codes) [iG1 7.Table 1]
[iG1
7.03]
|
Group
properties |
Three-
& Single-letter codes |
|
gly,ala,
val, leu,
ile, pro, met, phe, trp |
|
|
|
G
A V
L I
P M
F W |
|
gly, ser, thr, cys,
tyr, asn,
gln |
|
|
|
S
T
C Y
N Q |
|
lys, arg,
his |
|
|
|
K
R
H |
|
asp, glu |
|
|
|
D
E |
[Memorization
of
the abbreviations is not required for exams, but will
make your lives as biologists easier!]
in vitro
dehydration of
carboxy & amino termini forms peptide bond [iG1
7.05]

in vivo Peptidyl Transferase catalyzes
analogous condensation
reaction
carboxyl
(C) terminus of growing
polypeptide in P site
cleaved from tRNA &
joined to amino (N) terminus of new amino
acid in A site
NH2 - fmet - phe - gly - pro - COOH + NH2 - lys - COOH
which gives N - M F G P K - C
Repeating,
remnant backbone subunit [N - C(R) - C ] is amino
acid residue
Primary Structure - order
of amino acid residues in polypeptide
20N possible orders with N residues
Potential for enormous variety:
e.g., 205 = 3.2 x 106
possible pentapeptides
Secondary Structure - configuration of [-N-C(R)-C-]
backbone [iG1
6.06]
alpha helix:
a right-handed helix
beta-pleated-sheet: parallel
/ antiparallel chains
both
stabilized by H-bonds
Tertiary Structure - 3-Dimensional folding
of backbone [iG1 7.07]
Cys + Cys pairs form disulfide bridges
( - S - S -) [iG1
7.08]
Pro residues
form hydrophobic "corners"
hydrophilic residues
occur on exterior,
participate in reactions in aqueous environments
hydrophobic residues
occur in interior,
interact with membrane lipid bi-layer
Quaternary Structure - assembly of multiple
subunits [iG1 7.11]
monomers / dimers / oligomers
e.g., hemoglobin is a tetramer:
two alpha + two beta chains
charged residues (Asp, Glu, Lys, Arg, His)
form ionic bonds bx
subunits
Post-translational
processing (IG1 Table 8.6)
Chemical modification of
amino acids
addition of formyl
group to Met ![]() fMet
fMet
Addition
of carbohydrate side
chains (glycosylation) (IG1 8.21)
e.g., ABO blood group antigen proteins
Amino
acids may be cleaved out of primary structure (IG1 8.22,23,25)
e.g., biologically active insulin is less than half
the primary
sequence (IG1 8.24)
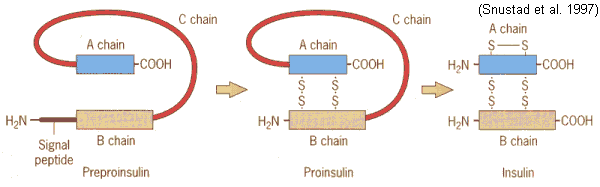
preproinsulin ![]() proinsulin
proinsulin ![]() insulin
insulin
(110 aa's)
(86 aa's)
(51 aa's)
Detection of genetically-based variation in Proteins
allelic variation translates to proteins with minor changes in electric charge
allozymes arise from different alleles of the same protein gene locus
substitution of alternatively charged amino acids changes overall charge
[cf: Isozymes are homologous proteins encoded by different gene loci]
Ex.: Sickle-cell hemoglobin (HbS) is a variant of standard HbA beta globin protein
Glu
"negative"
 net negative charge decreased
net negative charge decreasedVal stabilizes crystalline form of hemoglobin
 'sickling' of rbc's
Homework
#16: Critique the
following statement:
'sickling' of rbc's
Homework
#16: Critique the
following statement: "Electrophoresis of hemoglobin separates two alleles, F and S, of the Hb gene."
Enzymatic catalysis of biological reactions
Lowered energy of activation
biological reactions occur at body temperature
with lower energy input
Anabolic - synthesis of complex molecules from simpler components
Ex., transferases synthesize peptide bonds
synthetases attach tRNA to amino acid
Catabolic - break-down of complex molecules into simpler components
Ex., dehydrogenases remove protons (H+)
proteases split peptide bonds
Structural motifs recur in proteins with similar functions
Identification of motifs allows inferences about
function
Helix - turn - helix motifs binds Ca++ (IG1
RB9.1) (cf. iG1 7.12]
Zinc - finger motifs binds major &
minor DNA grooves (IG1 RB9.2)
Leucine Zipper motif binds
DNA and forms 'zippable' dimer (IG1 RB9.3)
Other protein functions
Structural
Collagen constitutes 25% of human protein
Histones are the major components of chromosomes
[online animation of
DNA packing into
chromosomes]
Nucleic Acid binding
Polymerases, nucleases, helicases, ligases, etc.
Transport
Hemoglobin in
blood & myoglobin in muscle bind O2
Drosophila Genome Project
has cataloged >11,000 genes
with protein products
>
1/2 have unknown functions
Human Genome comprises ~20,500 protein-coding genes: why
so few?
Protein-coding
regions may be transcribed in different
ways from different promoters
mRNAs may be spliced in different wasy
to combine different exons
All text material ©2015 by Steven M. Carr